Genome-Wide Identification of DNA Methyltransferase and Demethylase in Populus sect. Turanga and Their Potential Roles in Heteromorphic Leaf Development in Populus euphratica
- PMID: 40805719
- PMCID: PMC12349049
- DOI: 10.3390/plants14152370
Genome-Wide Identification of DNA Methyltransferase and Demethylase in Populus sect. Turanga and Their Potential Roles in Heteromorphic Leaf Development in Populus euphratica
Abstract
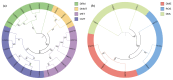
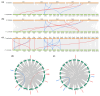
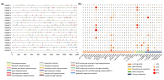
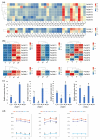
DNA methylation, mediated by DNA methyltransferases (DMTs) and demethylases (DMLs), is an important epigenetic modification that maintains genomic stability and regulates gene expression in plant growth, development, and stress responses. However, a comprehensive characterization of these gene families in Populus sect. Turanga remains lacking. In this study, eight PeDMT and two PeDML genes were identified in Populus euphratica, and six PpDMT and three PpDML genes in Populus pruinosa. Phylogenetic analysis revealed that DMTs and DMLs could be classified into four and three subfamilies, respectively. The analysis of cis-acting elements indicated that the promoter regions of both DMTs and DMLs were enriched with elements responsive to growth and development, light, phytohormones, and stress. Collinearity analysis detected three segmentally duplicated gene pairs (PeDMT5/8, PeDML1/2, and PpDML2/3), suggesting potential functional diversification. Transcriptome profiling showed that several PeDMTs and PeDMLs exhibited leaf shape- and developmental stage-specific expression patterns, with PeDML1 highly expressed during early stages and in broad-ovate leaves. Whole-genome bisulfite sequencing revealed corresponding decreases in DNA methylation levels, suggesting that active demethylation may contribute to heteromorphic leaf formation. Overall, this study provides significant insights for exploring the functions and expression regulation of plant DMTs and DMLs and will contribute to future research unraveling the molecular mechanisms of epigenetic regulation in P. euphratica.
Keywords: DML gene family; DMT gene famliy; Populus euphratica; Populus pruinosa; heteromorphic leaf.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


Similar articles
-
Transcriptome and Metabolome Analyses of Short-Term Responses of Populus talassica × Populus euphratica to Leaf Damage.Int J Mol Sci. 2025 Jun 19;26(12):5869. doi: 10.3390/ijms26125869. Int J Mol Sci. 2025. PMID: 40565333 Free PMC article.
-
Genome-Wide Identification and Expression Analysis of Aspartic proteases in Populus euphratica Reveals Candidates Involved in Salt Tolerance.Plants (Basel). 2025 Jun 23;14(13):1930. doi: 10.3390/plants14131930. Plants (Basel). 2025. PMID: 40647939 Free PMC article.
-
The Identification and Characterization of the PeGRF Gene Family in Populus euphratica Oliv. Heteromorphic Leaves Provide a Theoretical Basis for the Functional Study of PeGRF9.Int J Mol Sci. 2024 Dec 25;26(1):66. doi: 10.3390/ijms26010066. Int J Mol Sci. 2024. PMID: 39795925 Free PMC article.
-
Stage-specific DNA methylation dynamics in mammalian heart development.Epigenomics. 2025 Apr;17(5):359-371. doi: 10.1080/17501911.2025.2467024. Epub 2025 Feb 21. Epigenomics. 2025. PMID: 39980349 Review.
-
Identification of crucial genes sustaining the curled leaf phenotype in perennial transgenic poplar via advanced proteomic and phosphoproteomic analyses.J Proteomics. 2025 Aug 15;319:105471. doi: 10.1016/j.jprot.2025.105471. Epub 2025 Jun 3. J Proteomics. 2025. PMID: 40472892 Review.
References
Grants and funding
- 32371838/Natural Science Foundation of China
- 2023TSYCTD0019/Second batch of 'Tianshan' Support Program-Technology Innovation Team
- 2023ZD091, 2024ZD087/Guidance Program for Science & Technology Projects of XPCC
- TDZKBS202532/Start-up Research Fund for Doctoral Scholars from the President's Fund of Tarim University
LinkOut - more resources
Full Text Sources

