Drought Modulates Root-Microbe Interactions and Functional Gene Expression in Plateau Wetland Herbaceous Plants
- PMID: 40805763
- PMCID: PMC12349268
- DOI: 10.3390/plants14152413
Drought Modulates Root-Microbe Interactions and Functional Gene Expression in Plateau Wetland Herbaceous Plants
Abstract
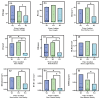
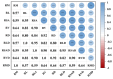
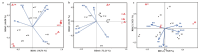
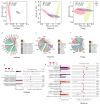
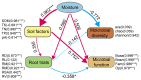
In plateau wetlands, the interactions of herbaceous roots with ectorhizosphere soil microorganisms represent an important way to realize their ecological functions. Global change-induced aridification of plateau wetlands has altered long-established functional synergistic relationships between plant roots and ectorhizosphere soil microbes, but we still know little about this phenomenon. In this context, nine typical wetlands with three different moisture statuses were selected from the eastern Tibetan Plateau in this study to analyze the relationships among herbaceous plant root traits and microbial communities and functions. The results revealed that drought significantly inhibited the accumulation of root biomass and surface area as well as the development of root volumes and diameters. Similarly, drought significantly reduced the diversity of ectorhizosphere soil microbial communities and the relative abundances of key phyla of archaea and bacteria. Redundancy analysis revealed that plant root traits and ectorhizosphere soil microbes were equally regulated by soil physicochemical properties. Functional genes related to carbohydrate metabolism were significantly associated with functional traits related to plant root elongation and nutrient uptake. Functional genes related to carbon and energy metabolism were significantly associated with traits related to plant root support and storage. Key genes such as CS,gltA, and G6PD,zwf help to improve the drought resistance and barrenness resistance of plant roots. This study helps to elucidate the synergistic mechanism of plant and soil microbial functions in plateau wetlands under drought stress, and provides a basis for evolutionary research and conservation of wetland ecosystems in the context of global change.
Keywords: functional genes; moisture status; plant root traits; plant–microbe interactions.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



Similar articles
-
Environment selected microbial function rather than taxonomic species in a plateau saline-alkaline wetland.Appl Environ Microbiol. 2025 Jul 23;91(7):e0220624. doi: 10.1128/aem.02206-24. Epub 2025 Jul 3. Appl Environ Microbiol. 2025. PMID: 40607849 Free PMC article.
-
Rhizobacteria from vineyard and commercial arbuscular mycorrhizal fungi induce synergistic microbiome shifts within grapevine root systems.Sci Rep. 2025 Jul 30;15(1):27884. doi: 10.1038/s41598-025-12673-5. Sci Rep. 2025. PMID: 40739286 Free PMC article.
-
Plant-soil feedback responses to drought are species-specific and only marginally predicted by root traits.Plant Soil. 2025;511(1-2):1205-1220. doi: 10.1007/s11104-024-07049-z. Epub 2024 Nov 7. Plant Soil. 2025. PMID: 40621105 Free PMC article.
-
Continuous cropping obstacles in medicinal plants: Driven by soil microbial communities and root exudates. A review.Plant Sci. 2025 Oct;359:112686. doi: 10.1016/j.plantsci.2025.112686. Epub 2025 Jul 26. Plant Sci. 2025. PMID: 40721126 Review.
-
The impact of aminoglycoside exposure on soil and plant root-associated microbiota: a meta-analysis.Environ Evid. 2025 Jul 10;14(1):12. doi: 10.1186/s13750-025-00365-6. Environ Evid. 2025. PMID: 40640967 Free PMC article.
References
-
- Chen Y., Lyu X. Research Directions in Wetland Function and Wetland Science. Wetl. Sci. 2003;5:7–11.
-
- Zhao Z., Zhang Y., Liu L., Liu F., Zhang H. Recent changes in wetlands on the Tibetan Plateau: A review. J. Geogr. Sci. 2015;25:879–896. doi: 10.1007/s11442-015-1208-5. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

