Genome-Wide Identification, Evolution, and Expression Patterns of the Fructose-1,6-Bisphosphatase Gene Family in Saccharum Species
- PMID: 40805782
- PMCID: PMC12349412
- DOI: 10.3390/plants14152433
Genome-Wide Identification, Evolution, and Expression Patterns of the Fructose-1,6-Bisphosphatase Gene Family in Saccharum Species
Abstract
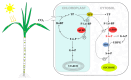
Fructose-1,6-bisphosphatase (FBP) is a crucial regulatory enzyme in sucrose synthesis and photosynthetic carbon assimilation, functioning through two distinct isoforms: cytosolic FBP (cyFBP) and chloroplastic FBP (cpFBP). However, the identification and functional characterization of FBP genes in Saccharum remains limited. In this study, we conducted a systematic identification and comparative genomics analyses of FBPs in three Saccharum species. We further examined their expression patterns across leaf developmental zones, spatiotemporal profiles, and responses to diurnal rhythms and hormonal treatments. Our analysis identified 95 FBP genes, including 44 cyFBPs and 51 cpFBPs. Comparative analyses revealed significant divergence in physicochemical properties, gene structures, and motif compositions between the two isoforms. Expression profiling indicated that both cyFBPs and cpFBPs were predominantly expressed in leaves, particularly in maturing and mature zones. During diurnal cycles, their expression peaked around the night-day transition, with cpFBPs exhibiting earlier peaks than cyFBPs. FBP genes in Saccharum spontaneum displayed greater diurnal sensitivity than those in Saccharum officinarum. Hormonal treatments further revealed significant regulatory divergence in FBP genes, both between isoforms and across species. Notably, cyFBP_2 and cpFBP_2 members consistently exhibited higher expression levels across all datasets, suggesting their pivotal roles in sugarcane physiology. These findings not only identify potential target genes for enhancing sucrose accumulation, but also highlight the breeding value of S. spontaneum and S. officinarum in sugarcane breeding.
Keywords: FBP gene family; Saccharum officinarum; Saccharum spontaneum; expression patterns; functional divergence; genome-wide identification.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures









Similar articles
-
Identification and characterization of PAL genes involved in the regulation of stem development in Saccharum spontaneum L.BMC Genom Data. 2024 Apr 30;25(1):38. doi: 10.1186/s12863-024-01219-9. BMC Genom Data. 2024. PMID: 38689211 Free PMC article.
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
[Identification of the sugarcane β-1,3-glucanase gene family and analysis of their expression under various stress conditions].Sheng Wu Gong Cheng Xue Bao. 2025 Jul 25;41(7):2913-2933. doi: 10.13345/j.cjb.250001. Sheng Wu Gong Cheng Xue Bao. 2025. PMID: 40769569 Chinese.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2017 Dec 22;12(12):CD011535. doi: 10.1002/14651858.CD011535.pub2. Cochrane Database Syst Rev. 2017. Update in: Cochrane Database Syst Rev. 2020 Jan 9;1:CD011535. doi: 10.1002/14651858.CD011535.pub3. PMID: 29271481 Free PMC article. Updated.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
References
-
- Koßmann J., Müller-Röber B., Dyer T.A., Raines C.A., Sonnewald U., Willmitzer L. Cloning and expression analysis of the plastidic fructose-l,6-bisphosphatase coding sequence from potato: Circumstantial evidence for the import of hexoses into chloroplasts. Planta. 1992;188:7–12. doi: 10.1007/BF01160706. - DOI - PubMed
Grants and funding
- (2022YFD2301101/National Key Research and Development 636 Program of China
- NKLTCB-YAAS-2024-S09, SKLTCB-YAAS-2025-07)/National Key Laboratory for Tropical Crop Breeding
- HHZ202201/Yunnan Haizhi Station for Sugarcane 638 Research Institute of Yunnan Academy of Agricultural Sciences
- 202205AM070001/Yunnan Science 639 and Technology Talent and platform program
- CARS-17/The Earmarked Fund for China 640 Agriculture Research System
LinkOut - more resources
Full Text Sources
Miscellaneous

