Screening of qPCR Reference Genes in Quinoa Under Cold, Heat, and Drought Gradient Stress
- PMID: 40805783
- PMCID: PMC12349563
- DOI: 10.3390/plants14152434
Screening of qPCR Reference Genes in Quinoa Under Cold, Heat, and Drought Gradient Stress
Abstract
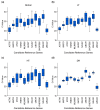
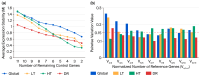
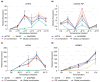
Quinoa (Chenopodium quinoa), a stress-tolerant pseudocereal ideal for studying abiotic stress responses, was used to systematically identify optimal reference genes for qPCR normalization under gradient stresses: low temperatures (LT group: -2 °C to -10 °C), heat (HT group: 39° C to 45 °C), and drought (DR group: 7 to 13 days). Through multi-algorithm evaluation (GeNorm, NormFinder, BestKeeper, the ΔCt method, and RefFinder) of eleven candidates, condition-specific optimal genes were established as ACT16 (Actin), SAL92 (IT4 phosphatase-associated protein), SSU32 (Ssu72-like family protein), and TSB05 (Tryptophan synthase beta-subunit 2) for the LT group; ACT16 and NRP13 (Asparagine-rich protein) for the HT group; and ACT16, SKP27 (S-phase kinase), and NRP13 for the DR group, with ACT16, NRP13, WLIM96 (LIM domain-containing protein), SSU32, SKP27, SAL92, and UBC22 (ubiquitin-conjugating enzyme E2) demonstrating cross-stress stability (global group). DHDPS96 (dihydrodipicolinate synthase) and EF03 (translation elongation factor) showed minimal stability. Validation using stress-responsive markers-COR72 (LT), HSP44 (HT), COR413-PM (LT), and DREB12 (DR)-confirmed reliability; COR72 and COR413-PM exhibited oscillatory cold response patterns, HSP44 peaked at 43 °C before declining, and DREB12 showed progressive drought-induced upregulation. Crucially, normalization with unstable genes (DHDPS96 and EF03) distorted expression profiles. This work provides validated reference standards for quinoa transcriptomics under abiotic stresses.
Keywords: Chenopodium quinoa; RT-qPCR; drought stress; expression stability; gradient stress; heat stress; low-temperature stress; reference genes.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


Similar articles
-
Deciphering melatonin biosynthesis pathway in Chenopodium quinoa: genome-wide analysis and expression levels of the genes under salt and drought.Planta. 2025 Jun 12;262(1):23. doi: 10.1007/s00425-025-04741-x. Planta. 2025. PMID: 40504264 Free PMC article.
-
Selection and Validation of Reference Genes for RT-qPCR in Protonemal Tissue of the Desiccation-Tolerant Moss Pseudocrossidium replicatum Under Multiple Abiotic Stress Conditions.Plants (Basel). 2025 Jun 7;14(12):1752. doi: 10.3390/plants14121752. Plants (Basel). 2025. PMID: 40573739 Free PMC article.
-
Development and selection of stably expressed reference genes for expression normalization in Ribes odoratum under drought stress.Sci Rep. 2025 Aug 9;15(1):29214. doi: 10.1038/s41598-025-12051-1. Sci Rep. 2025. PMID: 40783412 Free PMC article.
-
Validation of reference genes for RT-qPCR in marine bivalve ecotoxicology: Systematic review and case study using copper treated primary Ruditapes philippinarum hemocytes.Aquat Toxicol. 2017 Apr;185:86-94. doi: 10.1016/j.aquatox.2017.01.003. Epub 2017 Jan 16. Aquat Toxicol. 2017. PMID: 28189915
-
Thermal stability and storage of human insulin.Cochrane Database Syst Rev. 2023 Nov 6;11(11):CD015385. doi: 10.1002/14651858.CD015385.pub2. Cochrane Database Syst Rev. 2023. PMID: 37930742 Free PMC article.
References
-
- Ghadirnezhad S.S.R., Rahimi R., Zand-Silakhoor A., Fathi A., Fazeli A., Radicetti E., Mancinelli R. Enhancing seed germination under abiotic stress: Exploring the potential of nano-fertilization. J. Soil Sci. Plant Nutr. 2024;24:5319–5341. doi: 10.1007/s42729-024-01910-x. - DOI
-
- Alvar-Beltrán J., Verdi L., Marta A.D., Dao A., Vivoli R., Sanou J., Orlandini S. The effect of heat stress on quinoa (cv. Titicaca) under controlled climatic conditions. J. Agric. Sci. 2020;158:255–261. doi: 10.1017/S0021859620000556. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

