Identifying Therapeutic Targets for Amyotrophic Lateral Sclerosis Through Modeling of Multi-Omics Data
- PMID: 40806220
- PMCID: PMC12346086
- DOI: 10.3390/ijms26157087
Identifying Therapeutic Targets for Amyotrophic Lateral Sclerosis Through Modeling of Multi-Omics Data
Abstract
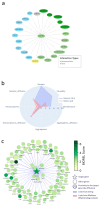
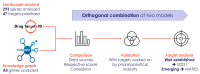
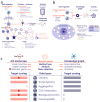
Amyotrophic lateral sclerosis (ALS) is a progressive neurodegenerative disease that primarily affects motor neurons, leading to loss of muscle control, and, ultimately, respiratory failure and death. Despite some advances in recent years, the underlying genetic and molecular mechanisms of ALS remain largely elusive. In this respect, a better understanding of these mechanisms is needed to identify new and biologically relevant therapeutic targets that could be developed into treatments that are truly disease-modifying, in that they address the underlying causes rather than the symptoms of ALS. In this study, we used two approaches to model multi-omics data in order to map and elucidate the genetic and molecular mechanisms involved in ALS, i.e., the molecular landscape building approach and the Patrimony platform. These two methods are complementary because they rely upon different omics data sets, analytic methods, and scoring systems to identify and rank therapeutic target candidates. The orthogonal combination of the two modeling approaches led to significant convergences, as well as some complementarity, both for validating existing therapeutic targets and identifying novel targets. As for validating existing targets, we found that, out of 217 different targets that have been or are being investigated for drug development, 10 have high scores in both the landscape and Patrimony models, suggesting that they are highly relevant for ALS. Moreover, through both models, we identified or corroborated novel putative drug targets for ALS. A notable example of such a target is MATR3, a protein that has strong genetic, molecular, and functional links with ALS pathology. In conclusion, by using two distinct and highly complementary disease modeling approaches, this study enhances our understanding of ALS pathogenesis and provides a framework for prioritizing new therapeutic targets. Moreover, our findings underscore the potential of leveraging multi-omics analyses to improve target discovery and accelerate the development of effective treatments for ALS, and potentially other related complex human diseases.
Keywords: MATR3; amyotrophic lateral sclerosis (ALS); molecular landscape; multi-omics modeling; patrimony; therapeutic targets.
Conflict of interest statement
Authors François Xavier Blaudin de Thé, Philippe Delagrange, Clotilde Mannoury La Cour, Mélanie Fouesnard, Sahar Elouej, Keith Mayl, Nicolas Lévy, Johannes Krupp, Ross Jeggo, and Philippe Moingeon were employed by the company Servier. Author Geert Poelmans was employed by the company Drug Target ID, Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

