Fever in Children with Cancer: Pathophysiological Insights Using Blood Transcriptomics
- PMID: 40806258
- PMCID: PMC12346811
- DOI: 10.3390/ijms26157126
Fever in Children with Cancer: Pathophysiological Insights Using Blood Transcriptomics
Abstract
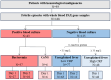
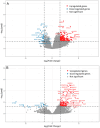
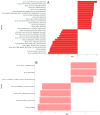
Fever is a frequent complication in children receiving chemotherapy, primarily caused by bloodstream infections and non-infectious inflammation. Yet, the pathophysiological mechanisms remain unclear, and diagnostics are insufficient, which often results in continued antibiotic treatment despite negative blood cultures. In a nationwide study, we collected whole blood in PAXgene tubes from 168 febrile episodes in children with hematological malignancies, including 37 episodes with bacteremia, and performed single-cell RNA sequencing. We compared transcriptomic profiles between febrile children with and without bacteremia. In children with bacteremia, differentially expressed genes were related to immunoregulation and cardiac and vascular function. Children without bacteremia had distinct gene expression patterns, suggesting a viral or other inflammatory cause of fever. Several differentially expressed genes overlapped with previously published transcriptomics-based diagnostic signatures developed in immunocompetent children. In conclusion, blood transcriptomics provided novel insights into the pathophysiological mechanisms of febrile children with hematological malignancies. We found differentially expressed genes suggesting viral infections or non-bacterial inflammation as causes of fever in children with negative blood cultures, supporting early antibiotic discontinuation in children with cancer.
Keywords: bloodstream infection; differential gene expression; febrile neutropenia; pediatric oncology; transcriptomics.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Castagnola E., Fontana V., Caviglia I., Caruso S., Faraci M., Fioredda F., Garrè M.L., Moroni C., Conte M., Losurdo G., et al. A Prospective Study on the Epidemiology of Febrile Episodes during Chemotherapy-Induced Neutropenia in Children with Cancer or after Hemopoietic Stem Cell Transplantation. Clin. Infect. Dis. 2007;45:1296–1304. doi: 10.1086/522533. - DOI - PubMed
-
- van der Velden F.J.S., de Vries G., Martin A., Lim E., von Both U., Kolberg L., Carrol E.D., Khanijau A., Herberg J.A., De T., et al. Febrile illness in high-risk children: A prospective, international observational study. Eur. J. Pediatr. 2023;182:543–554. doi: 10.1007/s00431-022-04642-1. - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

