Lupeol Attenuates Oxysterol-Induced Dendritic Cell Activation Through NRF2-Mediated Antioxidant and Anti-Inflammatory Effects
- PMID: 40806311
- PMCID: PMC12346829
- DOI: 10.3390/ijms26157179
Lupeol Attenuates Oxysterol-Induced Dendritic Cell Activation Through NRF2-Mediated Antioxidant and Anti-Inflammatory Effects
Abstract
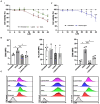
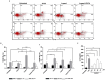
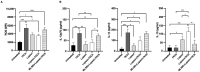
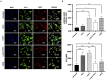
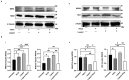
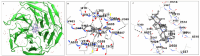
Oxysterols such as 7-ketocholesterol (7KCh) contribute to the pathogenesis of autoimmune and chronic inflammatory diseases by inducing oxidative stress and promoting pro-inflammatory immune cell activation. Dendritic cells (DCs) play a central role in maintaining immune tolerance, and their dysregulation is a key driver of autoimmunity. Targeting DCs by using natural compounds offers a promising strategy to restore redox balance and suppress aberrant immune responses. This study investigated the immunomodulatory and antioxidant properties of Lupeol, a natural triterpenoid, in human monocyte-derived DCs exposed to 7KCh. Flow cytometry and cytokine profiling demonstrated that Lupeol preserved the immature, tolerogenic phenotype of DCs by promoting a dose-dependent increase in the anti-inflammatory cytokine IL-10. Lupeol also inhibited the 7KCh-induced upregulation of maturation markers (CD83, CD86) and suppressed the release of pro-inflammatory cytokines IL-1β and IL-12p70. Functionally, Lupeol-treated DCs directed T cell polarization toward an anti-inflammatory and regulatory profile while dampening the inflammatory responses triggered by 7KCh. This immunoregulatory effect was further supported by the decreased secretion of the pro-inflammatory cytokines IL-1β and IL-12p70 in DC culture supernatants. Mechanistic analyses using immunofluorescence showed that Lupeol alone significantly increased nuclear NRF2 levels and upregulated HO-1 expression. Western blot analysis further confirmed Lupeol's ability to activate the KEAP1-NRF2 signaling pathway, as evidenced by increased expression of NRF2 and its downstream target, NQO1. The use of ML385, a selective NRF2 inhibitor, in ROS and cytokine assays supported the involvement of NRF2 in mediating the Lupeol antioxidant and anti-inflammatory effects in DCs. Notably, the oxidative burden induced by 7KCh limited the full activation of NRF2 signaling triggered by Lupeol. Furthermore, docking and MM/PBSA analyses revealed the specific interactions of Lupeol with the kelch domain of KEAP1. These findings suggest that Lupeol may serve as a promising orally available immunomodulatory agent capable of promoting tolerogenic DCs, offering potential applications in autoimmune and other chronic inflammatory diseases.
Keywords: autoimmune diseases; dendritic cells; natural compound.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
Astragaloside IV Attenuates Chronic Prostatitis by Activating Keap1/Nrf2/HO-1 Pathway: Suppressing Ferroptosis and Enhancing Antioxidant Defense.J Inflamm Res. 2025 Jul 29;18:10061-10079. doi: 10.2147/JIR.S527722. eCollection 2025. J Inflamm Res. 2025. PMID: 40761383 Free PMC article.
-
Narirutin mitigates inflammatory arthritis and osteoporosis through modulating macrophage phenotype and osteoclastogenesis.J Orthop Translat. 2025 Aug 8;54:115-130. doi: 10.1016/j.jot.2025.07.008. eCollection 2025 Sep. J Orthop Translat. 2025. PMID: 40822517 Free PMC article.
-
Management of urinary stones by experts in stone disease (ESD 2025).Arch Ital Urol Androl. 2025 Jun 30;97(2):14085. doi: 10.4081/aiua.2025.14085. Epub 2025 Jun 30. Arch Ital Urol Androl. 2025. PMID: 40583613 Review.
-
Microbiota-dependent modulation of intestinal anti-inflammatory CD4+ T cell responses.Semin Immunopathol. 2025 Apr 1;47(1):23. doi: 10.1007/s00281-025-01049-6. Semin Immunopathol. 2025. PMID: 40167791 Review.
References
LinkOut - more resources
Full Text Sources
Miscellaneous

