Single-Cell Dissection of the Serrated Pathway: Cellular Heterogeneity and Genetic Causality in Colorectal Cancer
- PMID: 40806320
- PMCID: PMC12347469
- DOI: 10.3390/ijms26157187
Single-Cell Dissection of the Serrated Pathway: Cellular Heterogeneity and Genetic Causality in Colorectal Cancer
Abstract
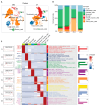
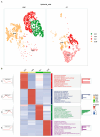
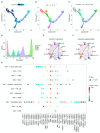
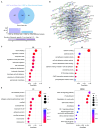
The serrated pathway represents a significant route to colorectal cancer (CRC), accounting for approximately 15-30% of cases, yet the specific epithelial cell subpopulations driving this pathway remain poorly understood. This study explores the causal relationship between serrated epithelial cells and CRC risk using single-cell transcriptomics and Mendelian randomization (MR). Publicly available single-cell RNA sequencing data were utilized to analyze epithelial cell subpopulations in CRC, focusing on specific serrated cells (SSCs). By integrating genome-wide association study data, MR was employed to assess the causal relationship between gene expression patterns and CRC risk. The study found that an increase in SSCs is closely associated with CRC progression. MR analysis revealed a significant correlation between expression changes in specific genes, such as IER3 in SSCs, and CRC risk (p < 0.05). Functional analyses indicated that IER3 may promote malignancy by regulating cell proliferation, adhesion, and immune evasion. Several genetic loci related to SSC gene expression were identified and validated for CRC risk association. This study demonstrates the significant role of serrated epithelial cell subpopulations in CRC development, particularly through key genes such as IER3, providing new perspectives for understanding CRC pathogenesis and future therapeutic strategies.
Keywords: Mendelian randomization; colorectal cancer; serrated epithelial cell; single-cell transcriptomics; tumor progression.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflicts of interest.
Figures


References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

