Dysregulated miRNAs Targeting Adiponectin Signaling in Colorectal Cancer
- PMID: 40806335
- PMCID: PMC12346623
- DOI: 10.3390/ijms26157196
Dysregulated miRNAs Targeting Adiponectin Signaling in Colorectal Cancer
Abstract
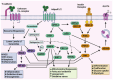
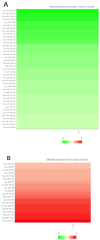
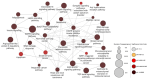
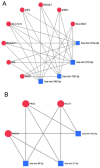
Dysregulation in miRNA expression has been reported in a variety of tumors, including colorectal cancer (CRC), where adiponectin regulates a number of processes related to tumorigenesis. The aim of this study was to identify a panel of heavily and consistently altered miRNAs in CRC that affect adiponectin signaling based on bioinformatics analysis and cross-referencing the available literature. Bioinformatics tools were used to analyze publicly available datasets to identify miRNAs targeting the adiponectin pathway that are substantially dysregulated in CRC. In parallel, a comprehensive literature review was conducted to gather and explore existing knowledge on the relationship between CRC, adiponectin signaling, and miRNA dysregulation. Bioinformatics analysis revealed a set of miRNAs that target adiponectin signaling and are consistently altered in CRC. Several candidate miRNAs, including miR-215-5p, miR-340-5p, miR-181a-5p, miR-150-5p, miR-96-5p, miR-19a-3p, and miR-21-5p, were identified as potential key regulators of the adiponectin cascade, while also being systemically dysregulated in CRC. Through gene ontology enrichment analysis, we further elucidated the biological processes and pathways impacted by these miRNAs, providing insight into their contributions to CRC. The literature review did not identify any previously reported shared connection between these miRNAs, adiponectin signaling, and CRC pathogenesis.
Keywords: adiponectin signaling; colorectal cancer; miRNAs.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
-
- Mehterov N., Sacconi A., Pulito C., Vladimirov B., Haralanov G., Pazardjikliev D., Nonchev B., Berindan-Neagoe I., Blandino G., Sarafian V. A novel panel of clinically relevant miRNAs signature accurately differentiates oral cancer from normal mucosa. Front. Oncol. 2022;12:1072579. doi: 10.3389/fonc.2022.1072579. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

