Development of a Broad-Spectrum Pan-Mpox Vaccine via Immunoinformatic Approaches
- PMID: 40806344
- PMCID: PMC12346078
- DOI: 10.3390/ijms26157210
Development of a Broad-Spectrum Pan-Mpox Vaccine via Immunoinformatic Approaches
Abstract
Monkeypox virus (MPXV) has caused 148,892 confirmed cases and 341 deaths from 137 countries worldwide, as reported by the World Health Organization (WHO), highlighting the urgent need for effective vaccines to prevent the spread of MPXV. Traditional vaccine development is low-throughput, expensive, time consuming, and susceptible to reversion to virulence. Alternatively, a reverse vaccinology approach offers a rapid, efficient, and safer alternative for MPXV vaccine design. Here, MPXV proteins associated with viral infection were analyzed for immunogenic epitopes to design multi-epitope vaccines based on B-cell, CD4+, and CD8+ epitopes. Epitopes were selected based on allergenicity, antigenicity, and toxicity parameters. The prioritized epitopes were then combined via peptide linkers and N-terminally fused to various protein adjuvants, including PADRE, beta-defensin 3, 50S ribosomal protein L7/12, RS-09, and the cholera toxin B subunit (CTB). All vaccine constructs were computationally validated for physicochemical properties, antigenicity, allergenicity, safety, solubility, and structural stability. The three-dimensional structure of the selected construct was also predicted. Moreover, molecular docking and molecular dynamics (MD) simulations between the vaccine and the TLR-4 immune receptor demonstrated a strong and stable interaction. The vaccine construct was codon-optimized for high expression in the E. coli and was finally cloned in silico into the pET21a (+) vector. Collectively, these results could represent innovative tools for vaccine formulation against MPXV and be transformative for other infectious diseases.
Keywords: Mpox; broad-spectrum vaccine; immunoinformatics approach; molecular docking; molecular dynamics (MD) simulations.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

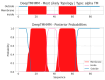

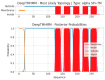
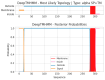
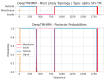










Similar articles
-
Immunoinformatic design of chimeric multiepitope vaccine for the prevention of human metapneumovirus (hMPV).BMC Infect Dis. 2025 Jul 30;25(1):964. doi: 10.1186/s12879-025-11339-x. BMC Infect Dis. 2025. PMID: 40739488 Free PMC article.
-
Immuno-informatics analyses of important esophageal cancer associated viruses for multi-epitope vaccine design.Front Immunol. 2025 Jul 8;16:1587224. doi: 10.3389/fimmu.2025.1587224. eCollection 2025. Front Immunol. 2025. PMID: 40698087 Free PMC article.
-
Next-generation multi-epitope subunit vaccine design: A computational approach utilizing two stable proteins to combat Human Metapneumovirus (HMPV).Comput Biol Med. 2025 Sep;196(Pt C):110935. doi: 10.1016/j.compbiomed.2025.110935. Epub 2025 Aug 18. Comput Biol Med. 2025. PMID: 40829349
-
Immunoinformatics-Driven Design of a Multi-Epitope Vaccine Targeting Simian Virus VP1 Major Capsid Protein for Oncogenic Viral Infection Prevention.Rev Med Virol. 2025 Sep;35(5):e70065. doi: 10.1002/rmv.70065. Rev Med Virol. 2025. PMID: 40781525 Review.
-
Multi-epitope vaccines: charting a new frontier in monkeypox prevention and control.Hum Cell. 2025 Jul 9;38(5):126. doi: 10.1007/s13577-025-01255-2. Hum Cell. 2025. PMID: 40632349 Review.
References
-
- Yang Z.-S., Lin C.-Y., Urbina A.N., Wang W.-H., Assavalapsakul W., Tseng S.-P., Lu P.-L., Chen Y.-H., Yu M.-L., Wang S.-F. The First Case of Monkeypox Virus Infection Detected in Taiwan: Awareness and Preparation. Int. J. Infect. Dis. 2022;122:991–995. doi: 10.1016/j.ijid.2022.07.051. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

