Physiological and Transcriptomic Analyses Reveal Regulatory Mechanisms of Adventitious Root Formation in In Vitro Culture of Cinnamomum camphora
- PMID: 40806399
- PMCID: PMC12347477
- DOI: 10.3390/ijms26157264
Physiological and Transcriptomic Analyses Reveal Regulatory Mechanisms of Adventitious Root Formation in In Vitro Culture of Cinnamomum camphora
Abstract
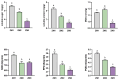
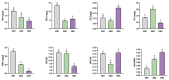
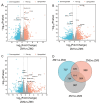
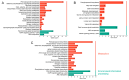
Cinnamomum camphora is an ecologically and economically significant species, highly valued for its essential oil production and environmental benefits. Although a tissue culture system has been established for C. camphora, large-scale propagation remains limited due to the inconsistent formation of adventitious roots (ARs). This study investigated AR formation from callus tissue, focusing on associated physiological changes and gene expression dynamics. During AR induction, contents of soluble sugars and proteins decreased, alongside reduced activities of antioxidant enzymes, including superoxide dismutase (SOD), peroxidase (POD), and polyphenol oxidase (PPO). Levels of indole-3-acetic acid (IAA) and abscisic acid (ABA) decreased significantly throughout AR formation. Zeatin riboside (ZR) levels initially declined and then rose, whereas gibberellic acid (GA) levels displayed the opposite trend. Comparative transcriptomic and temporal expression analyses identified differentially expressed genes (DEGs), which were grouped into four distinct expression patterns. KEGG pathway enrichment indicated that 67 DEGs are involved in plant hormone signaling pathways and that 38 DEGs are involved in the starch and sucrose metabolism pathway. Additionally, protein-protein interaction network (PPI) analysis revealed ten key regulatory genes, which are mainly involved in auxin, cytokinin, GA, ABA, and ethylene signaling pathways. The reliability of the transcriptome data was further validated by quantitative real-time PCR. Overall, this study provides new insights into the physiological and molecular mechanisms underlying AR formation in C. camphora and offers valuable guidance for optimizing tissue culture systems.
Keywords: RNA-seq; endogenous hormones; morphogenesis; oxidoreductases; temporal dynamics.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures






Similar articles
-
Transcriptome analysis reveals rootstock-driven effects on growth and photosynthesis in Camellia chekiangoleosa: A phenotypic and biochemical perspective.PLoS One. 2025 Sep 3;20(9):e0331313. doi: 10.1371/journal.pone.0331313. eCollection 2025. PLoS One. 2025. PMID: 40901902 Free PMC article.
-
Regulatory roles of indole-3-acetic acid in the physiological and biochemical responses of Cinnamomum camphora seedlings under cadmium stress.Ecotoxicol Environ Saf. 2025 Sep 1;302:118671. doi: 10.1016/j.ecoenv.2025.118671. Epub 2025 Jul 15. Ecotoxicol Environ Saf. 2025. PMID: 40663943
-
Multiomics analysis of gibberellin involved in far-red light-regulated internode elongation in cucumber seedlings.Plant Cell Rep. 2025 Sep 6;44(10):208. doi: 10.1007/s00299-025-03598-4. Plant Cell Rep. 2025. PMID: 40913742
-
To grow or not to grow: the enigma of plant root growth dynamism.Plant Mol Biol. 2025 Jul 30;115(4):93. doi: 10.1007/s11103-025-01631-4. Plant Mol Biol. 2025. PMID: 40736880 Review.
-
Adventitious Root Formation in Cuttings: Insights from Arabidopsis and Prospects for Woody Plants.Biomolecules. 2025 Jul 28;15(8):1089. doi: 10.3390/biom15081089. Biomolecules. 2025. PMID: 40867534 Free PMC article. Review.
References
-
- Takaoka D. Sesquiterpene alcohols in camphor oil. Phytochemistry. 1976;15:425–426. doi: 10.1016/S0031-9422(00)86839-9. - DOI
-
- Nirmal Babu K., Sajina A., Minoo D., John C.Z., Mini P.M., Tushar K.V., Rema J., Ravindran P.N. Micropropagation of camphor tree (Cinnamomum camphora) Plant Cell Tissue Organ Cult. 2003;74:179–183. doi: 10.1023/A:1023988110064. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

