The Human Mycobiome: Composition, Immune Interactions, and Impact on Disease
- PMID: 40806413
- PMCID: PMC12347658
- DOI: 10.3390/ijms26157281
The Human Mycobiome: Composition, Immune Interactions, and Impact on Disease
Abstract
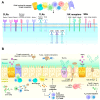
The fungal component of microbiota, known as the mycobiome, inhabits different body niches such as the skin and the gastrointestinal, respiratory, and genitourinary tracts. Much information has been gained on the bacterial component of the human microbiota, but the mycobiome has remained somewhat elusive due to its sparsity, variability, susceptibility to environmental factors (e.g., early life colonization, diet, or pharmacological treatments), and the specific in vitro culture challenges. Functionally, the mycobiome is known to play a role in modulating innate and adaptive immune responses by interacting with microorganisms and immune cells. The latter elicits anti-fungal responses via the recognition of specific fungal cell-wall components (e.g., β-1,3-glucan, mannan, and chitin) by immune system receptors. These receptors then regulate the activation and differentiation of many innate and adaptive immune cells including mucocutaneous cell barriers, macrophages, neutrophils, dendritic cells, natural killer cells, innate-like lymphoid cells, and T and B lymphocytes. Mycobiome disruptions have been correlated with various diseases affecting mostly the brain, lungs, liver and pancreas. This work reviews our current knowledge on the mycobiome, focusing on its composition, research challenges, conditioning factors, interactions with the bacteriome and the immune system, and the known mycobiome alterations associated with disease.
Keywords: autoimmunity; cancer; fungal receptors; immune cells; infection; inflammation; mycobiome.
Conflict of interest statement
F.L. is the founder and ad honorem scientific advisor of Sepsia Therapeutics S.L. M.I. is a founding partner of Sepsia Therapeutics S.L. The rest of the authors declare no conflicts of interest.
Figures
Similar articles
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
T-bet expressing Tr1 cells driven by dietary signals dominate the small intestinal immune landscape.bioRxiv [Preprint]. 2025 Jul 4:2025.06.30.662190. doi: 10.1101/2025.06.30.662190. bioRxiv. 2025. PMID: 40747421 Free PMC article. Preprint.
-
Type 1 Diabetes: A Guide to Autoimmune Mechanisms for Clinicians.Diabetes Obes Metab. 2025 Aug;27 Suppl 6(Suppl 6):40-56. doi: 10.1111/dom.16460. Epub 2025 May 15. Diabetes Obes Metab. 2025. PMID: 40375390 Free PMC article. Review.
-
Interactions Between the Innate and Adaptive Immune Responses.Adv Exp Med Biol. 2025;1476:297-308. doi: 10.1007/978-3-031-85340-1_12. Adv Exp Med Biol. 2025. PMID: 40622548 Review.
-
Management of urinary stones by experts in stone disease (ESD 2025).Arch Ital Urol Androl. 2025 Jun 30;97(2):14085. doi: 10.4081/aiua.2025.14085. Epub 2025 Jun 30. Arch Ital Urol Androl. 2025. PMID: 40583613 Review.
References
-
- Hallen-Adams H.E., Kachman S.D., Kim J., Legge R.M., Martínez I. Fungi Inhabiting the Healthy Human Gastrointestinal Tract: A Diverse and Dynamic Community. Fungal Ecol. 2015;15:9–17. doi: 10.1016/j.funeco.2015.01.006. - DOI
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

