Escalated Oxycodone Self-Administration Is Associated with Activation of Specific Gene Networks in the Rat Dorsal Striatum
- PMID: 40806484
- PMCID: PMC12347817
- DOI: 10.3390/ijms26157356
Escalated Oxycodone Self-Administration Is Associated with Activation of Specific Gene Networks in the Rat Dorsal Striatum
Abstract
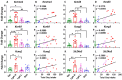
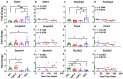
The diagnosis of opioid use disorder (OUD) is prevalent due to increased prescribing of opioids. Long-term oxycodone self-administration can lead to addiction-like behavioral responses in rats. Herein, we sought to identify molecular pathways consequent to long-term exposure to oxycodone self-administration. Towards that end, we used male Sprague Dawley rats that self-administered oxycodone for 20 days according to short-(ShA, 3 h) and long-access (LgA, 9 h) paradigms. LgA rats escalated their oxycodone intake and developed into 2 phenotypes, labeled Long-access High (LgA-H) and Long-access Low (LgA-L) rats, based on their escalation. RNA sequencing analysis revealed the LgA-H has significantly different DEGs in comparison to other groups. DAVID analysis revealed the participation of LgA-H DEGs in potassium transport. RT-PCR analysis of striatal samples validated the increased levels of potassium channels. Since these increases correlated with oxycodone intake, we believe potassium channels are potential targets for the treatment of oxycodone use disorder.
Keywords: RNA sequencing; dorsal striatum; oxycodone; potassium channels; self-administration.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
-
- National Institute on Drug Abuse Advancing Addiciton Science. [(accessed on 13 January 2025)]; Available online: https://nida.nih.gov/research-topics/trends-statistics/overdose-death-rates.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

