Transcriptomic Reprogramming and Key Molecular Pathways Underlying Huanglongbing Tolerance and Susceptibility in Six Citrus Cultivars
- PMID: 40806488
- PMCID: PMC12347598
- DOI: 10.3390/ijms26157359
Transcriptomic Reprogramming and Key Molecular Pathways Underlying Huanglongbing Tolerance and Susceptibility in Six Citrus Cultivars
Abstract
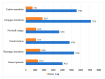
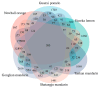
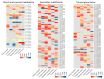
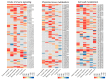
Huanglongbing (HLB), caused by Candidatus Liberibacter asiaticus (CLas), is the most devastating disease threatening global citrus production. Although no commercial citrus varieties exhibit complete HLB resistance, genotype-specific tolerance variations remain underexplored. This study conducted a comparative transcriptomic profiling of six commercially citrus cultivars in South China, four susceptible cultivars (C. reticulata cv. Tankan, Gongkan, Shatangju, and C. sinensis Osbeck cv. Newhall), and two tolerant cultivars (C. limon cv. Eureka; C. maxima cv Guanxi Yu) to dissect molecular mechanisms underlying HLB responses. Comparative transcriptomic analyses revealed extensive transcriptional reprogramming, with tolerant cultivars exhibiting fewer differentially expressed genes (DEGs) and targeted defense activation compared to susceptible genotypes. The key findings highlighted the genotype-specific regulation of starch metabolism, where β-amylase 3 (BAM3) was uniquely upregulated in tolerant varieties, potentially mitigating starch accumulation. Immune signaling diverged significantly: tolerant cultivars activated pattern-triggered immunity (PTI) via receptor-like kinases (FLS2) and suppressed ROS-associated RBOH genes, while susceptible genotypes showed the hyperactivation of ethylene signaling and oxidative stress pathways. Cell wall remodeling in susceptible cultivars involved upregulated xyloglucan endotransglucosylases (XTH), contrasting with pectin methylesterase induction in tolerant Eureka lemon for structural reinforcement. Phytohormonal dynamics revealed SA-mediated defense and NPR3/4 suppression in Eureka lemon, whereas susceptible cultivars prioritized ethylene/JA pathways. These findings delineate genotype-specific strategies in citrus-CLas interactions, identifying BAM3, FLS2, and cell wall modifiers as critical targets for breeding HLB-resistant cultivars through molecular-assisted selection. This study provides a foundational framework for understanding host-pathogen dynamics and advancing citrus immunity engineering.
Keywords: citrus cultivar; host response; huanglongbing; susceptible; tolerant.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
-
- Bové J.M. Huanglongbing: A destructive, newly-emerging, century-old disease of citrus. J. Plant Pathol. 2006;88:7–37.
-
- Teixeira D.D.C., Saillard C., Eveillard S., Danet J.L., Costa P.I.D., Ayres A.J., Bové J.M. ‘Candidatus Liberibacter americanus’, associated with citrus huanglongbing (greening disease) in São Paulo State, Brazil. Int. J. Syst. Evol. Microbiol. 2005;55:1857–1862. doi: 10.1099/ijs.0.63677-0. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

