Identification of Differentially Expressed Genes and Pathways in Non-Diabetic CKD and Diabetic CKD by Integrated Human Transcriptomic Bioinformatics Analysis
- PMID: 40806550
- PMCID: PMC12347806
- DOI: 10.3390/ijms26157421
Identification of Differentially Expressed Genes and Pathways in Non-Diabetic CKD and Diabetic CKD by Integrated Human Transcriptomic Bioinformatics Analysis
Abstract
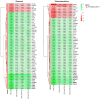
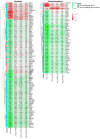
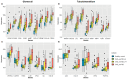
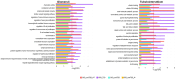
Chronic kidney disease (CKD) is a heterogeneous condition with various etiologies, including type 2 diabetes mellitus (T2D), hypertension, and autoimmune disorders. Both diabetic CKD (CKD_T2D) and non-diabetic CKD (CKD_nonT2D) share overlapping clinical features, but understanding the molecular mechanisms underlying each subtype and distinguishing diabetic from non-diabetic forms remain poorly defined. To identify differentially expressed genes (DEGs) and enriched biological pathways between CKD_T2D and CKD_nonT2D cohorts, including autoimmune (CKD_nonT2D_AI) and hypertensive (CKD_nonT2D_HT) subtypes, through integrative transcriptomic analysis. Publicly available gene expression datasets from human glomerular and tubulointerstitial kidney tissues were curated and analyzed from GEO and ArrayExpress. Differential expression analysis and Gene Set Enrichment Analysis (GSEA) were conducted to assess cohort-specific molecular signatures. A considerable overlap in DEGs was observed between CKD_T2D and CKD_nonT2D, with CKD_T2D exhibiting more extensive gene expression changes. Hypertensive-CKD shared greater transcriptomic similarity with CKD_T2D than autoimmune-CKD. Key DEGs involved in fibrosis, inflammation, and complement activation-including Tgfb1, Timp1, Cxcl6, and C1qa/B-were differentially regulated in diabetic samples, where GSEA revealed immune pathway enrichment in glomeruli and metabolic pathway enrichment in tubulointerstitium. The transcriptomic landscape of CKD_T2D reveals stronger immune and metabolic dysregulation compared to non-diabetic CKD. These findings suggest divergent pathological mechanisms and support the need for tailored therapeutic approaches.
Keywords: diabetic chronic kidney disease; differential expression analysis; gene set enrichment analysis; non-diabetic chronic kidney disease; transcriptomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Common molecular links and therapeutic insights between type 2 diabetes and kidney cancer.PLoS One. 2025 Aug 20;20(8):e0330619. doi: 10.1371/journal.pone.0330619. eCollection 2025. PLoS One. 2025. PMID: 40834023 Free PMC article.
-
Deciphering Shared Gene Signatures and Immune Infiltration Characteristics Between Gestational Diabetes Mellitus and Preeclampsia by Integrated Bioinformatics Analysis and Machine Learning.Reprod Sci. 2025 Jun;32(6):1886-1904. doi: 10.1007/s43032-025-01847-1. Epub 2025 May 15. Reprod Sci. 2025. PMID: 40374866
-
Integrated single-cell and transcriptomic analysis of bone marrow-derived metastatic neuroblastoma reveals molecular mechanisms of metabolic reprogramming.Sci Rep. 2025 Aug 5;15(1):28519. doi: 10.1038/s41598-025-13626-8. Sci Rep. 2025. PMID: 40764361 Free PMC article.
-
Altered dietary salt intake for preventing diabetic kidney disease and its progression.Cochrane Database Syst Rev. 2023 Jan 16;1(1):CD006763. doi: 10.1002/14651858.CD006763.pub3. Cochrane Database Syst Rev. 2023. PMID: 36645291 Free PMC article.
-
Early referral strategies for management of people with markers of renal disease: a systematic review of the evidence of clinical effectiveness, cost-effectiveness and economic analysis.Health Technol Assess. 2010 Apr;14(21):1-184. doi: 10.3310/hta14210. Health Technol Assess. 2010. PMID: 20441712
References
-
- Stevens P.E., Ahmed S.B., Carrero J.J., Foster B., Francis A., Hall R.K., Herrington W.G., Hill G., Inker L.A., Kazancıoğlu R., et al. KDIGO 2024 Clinical Practice Guideline for the Evaluation and Management of Chronic Kidney Disease. Kidney Int. 2024;105:S117–S314. doi: 10.1016/j.kint.2023.10.018. - DOI - PubMed
-
- Ortiz A., Covic A., Fliser D., Fouque D., Goldsmith D., Kanbay M., Mallamaci F., Massy Z.A., Rossignol P., Vanholder R., et al. Epidemiology, Contributors to, and Clinical Trials of Mortality Risk in Chronic Kidney Failure. Lancet. 2014;383:1831–1843. doi: 10.1016/S0140-6736(14)60384-6. - DOI - PubMed
-
- Obrador G.T., Schultheiss U.T., Kretzler M., Langham R.G., Nangaku M., Pecoits-Filho R., Pollock C., Rossert J., Correa-Rotter R., Stenvinkel P., et al. Genetic and Environmental Risk Factors for Chronic Kidney Disease. Kidney Int. Suppl. 2017;7:88–106. doi: 10.1016/j.kisu.2017.07.004. - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

