Supervised Machine-Based Learning and Computational Analysis to Reveal Unique Molecular Signatures Associated with Wound Healing and Fibrotic Outcomes to Lens Injury
- PMID: 40806551
- PMCID: PMC12347510
- DOI: 10.3390/ijms26157422
Supervised Machine-Based Learning and Computational Analysis to Reveal Unique Molecular Signatures Associated with Wound Healing and Fibrotic Outcomes to Lens Injury
Abstract
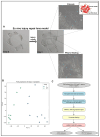
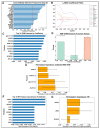
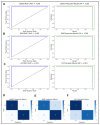
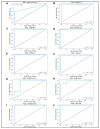
Posterior capsule opacification (PCO), a frequent complication of cataract surgery, arises from dysregulated wound healing and fibrotic transformation of residual lens epithelial cells. While transcriptomic and machine learning (ML) approaches have elucidated fibrosis-related pathways in other tissues, the molecular divergence between regenerative and fibrotic outcomes in the lens remains unclear. Here, we used an ex vivo chick lens injury model to simulate post-surgical conditions, collecting RNA from lenses undergoing either regenerative wound healing or fibrosis between days 1-3 post-injury. Bulk RNA sequencing data were normalized, log-transformed, and subjected to univariate filtering prior to training LASSO, SVM, and RF ML models to identify discriminatory gene signatures. Each model was independently validated using a held-out test set. Distinct gene sets were identified, including fibrosis-associated genes (VGLL3, CEBPD, MXRA7, LMNA, gga-miR-143, RF00072) and wound-healing-associated genes (HS3ST2, ID1), with several achieving perfect classification. Gene Set Enrichment Analysis revealed divergent pathway activation, including extracellular matrix remodeling, DNA replication, and spliceosome associated with fibrosis. RT-PCR in independent explants confirmed key differential expression levels. These findings demonstrate the utility of supervised ML for discovering lens-specific fibrotic and regenerative gene features and nominate biomarkers for targeted intervention to mitigate PCO.
Keywords: RNA sequencing; biomarker discovery; cataract surgery; fibrosis; lens; machine learning; posterior capsule opacification; repair; wound healing.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
Intraocular lens optic edge design for the prevention of posterior capsule opacification after cataract surgery.Cochrane Database Syst Rev. 2021 Aug 16;8(8):CD012516. doi: 10.1002/14651858.CD012516.pub2. Cochrane Database Syst Rev. 2021. PMID: 34398965 Free PMC article.
-
Trifocal versus extended depth of focus (EDOF) intraocular lenses after cataract extraction.Cochrane Database Syst Rev. 2024 Jul 10;7(7):CD014891. doi: 10.1002/14651858.CD014891.pub2. Cochrane Database Syst Rev. 2024. PMID: 38984608 Free PMC article.
-
Types of intraocular lenses for cataract surgery in eyes with uveitis.Cochrane Database Syst Rev. 2014 Mar 4;3(3):CD007284. doi: 10.1002/14651858.CD007284.pub2. Cochrane Database Syst Rev. 2014. PMID: 24590672 Free PMC article.
-
Transcriptomic Analysis of Human Lens Epithelium Tissue With and Without Cataract Surgery: Uncovering Novel Pathways of Post-Surgical Lens Epithelium Remodeling.Invest Ophthalmol Vis Sci. 2025 Jul 1;66(9):28. doi: 10.1167/iovs.66.9.28. Invest Ophthalmol Vis Sci. 2025. PMID: 40643341 Free PMC article.
-
Accommodative intraocular lens versus standard monofocal intraocular lens implantation in cataract surgery.Cochrane Database Syst Rev. 2014 May 1;2014(5):CD009667. doi: 10.1002/14651858.CD009667.pub2. Cochrane Database Syst Rev. 2014. PMID: 24788900 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

