PSG and Other Candidate Genes as Potential Biomarkers of Therapy Resistance in B-ALL: Insights from Chromosomal Microarray Analysis and Machine Learning
- PMID: 40806565
- PMCID: PMC12347871
- DOI: 10.3390/ijms26157437
PSG and Other Candidate Genes as Potential Biomarkers of Therapy Resistance in B-ALL: Insights from Chromosomal Microarray Analysis and Machine Learning
Abstract
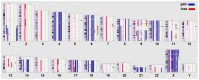
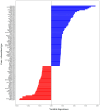
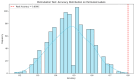
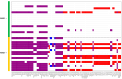
Chromosomal microarray analysis (CMA) was performed for 40 patients with B-ALL undergoing treatment according to the ALL-2016 protocol to investigate the copy number alterations (CNAs) and copy neutral loss of heterozygosity (cnLOH) associated with minimal residual disease (MRD)-positive remission. Aberrations involving over 20,000 genes were identified, and a random forest approach was applied to isolate a subset of genes whose CNAs and cnLOH are significantly associated with poor therapeutic response. We have assembled the triple matched healthy population data and used that data as a reference, but not as a matched control. We identified a recurrent cluster of cnLOH in the 19q13.2-19q13.31 region, significantly enriched in MRD-positive patients (70% vs. 47% in the reference group vs. 16% in MRD-negative patients). This region includes the pregnancy-specific glycoprotein (PSG) gene family and the oncogene ERF, suggesting a potential role in leukemic persistence and treatment resistance. Additionally, we observed significant deletions involving 7p22.3 and 16q13, often as part of large-scale losses affecting almost the entire chromosomes 7 and 16, indicative of global chromosomal instability. These findings highlight specific genomic regions potentially involved in therapy resistance and may contribute to improved risk stratification in B-ALL. Our findings emphasize the value of high-resolution CMA in diagnostics and risk stratification and suggest that PSG genes and other candidate genes could serve as biomarkers for predicting treatment outcomes.
Keywords: acute B-lymphoblastic leukemia (B-ALL); copy neutral loss of heterozygosity (cnLOH); copy number alterations (CNAs); genes; machine learning.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures






Similar articles
-
Can a Liquid Biopsy Detect Circulating Tumor DNA With Low-passage Whole-genome Sequencing in Patients With a Sarcoma? A Pilot Evaluation.Clin Orthop Relat Res. 2025 Jan 1;483(1):39-48. doi: 10.1097/CORR.0000000000003161. Epub 2024 Jun 21. Clin Orthop Relat Res. 2025. PMID: 38905450
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
Blinatumomab Therapy Is Associated with Favorable Outcomes after Allogeneic Hematopoietic Cell Transplantation in Pediatric Patients with B Cell Acute Lymphoblastic Leukemia.Transplant Cell Ther. 2024 Feb;30(2):217-227. doi: 10.1016/j.jtct.2023.10.024. Epub 2023 Nov 4. Transplant Cell Ther. 2024. PMID: 37931800
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
-
Impact of residual disease as a prognostic factor for survival in women with advanced epithelial ovarian cancer after primary surgery.Cochrane Database Syst Rev. 2022 Sep 26;9(9):CD015048. doi: 10.1002/14651858.CD015048.pub2. Cochrane Database Syst Rev. 2022. PMID: 36161421 Free PMC article.
References
-
- Parovichnikova E.N., Aleshina O.A., Troitskaya V.V., Chabaeva Y.A., Sokolov A.N., Isinova G.A., Kotova E.S., Akhmerzaeva Z.H., Klyasova G.A., Galtseva I.V., et al. Comparison of the treatment results in adult patients with acute Ph-negative lymphoblastic leukemia on protocols of the Russian multicenter studies ALL-2009 and ALL-2016. Russ. J. Hematol. Transfusiol. 2022;67:460–477. doi: 10.35754/0234-5730-2022-67-4-460-477. (In Russian) - DOI
-
- Miller D.T., Adam M.P., Aradhya S., Biesecker L.G., Brothman A.R., Carter N.P., Church D.M., Crolla J.A., Eichler E.E., Epstein C.J., et al. Consensus statement: Chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am. J. Hum. Genet. 2010;86:749–764. doi: 10.1016/j.ajhg.2010.04.006. - DOI - PMC - PubMed
-
- Kearney H.M., Thorland E.C., Brown K.K., Quintero-Rivera F., South S.T., Working Group of the American College of Medical Genetics Laboratory Quality Assurance Committee American College of Medical Genetics standards and guidelines for interpretation and reporting of postnatal constitutional copy number variants. Genet. Med. 2011;13:680–685. doi: 10.1097/GIM.0b013e3182217a3a. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

