Genome-Wide Analyses of the XTH Gene Family in Brachypodium distachyon and Functional Analyses of the Role of BdXTH27 in Root Elongation
- PMID: 40806582
- PMCID: PMC12347618
- DOI: 10.3390/ijms26157457
Genome-Wide Analyses of the XTH Gene Family in Brachypodium distachyon and Functional Analyses of the Role of BdXTH27 in Root Elongation
Abstract
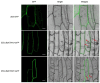
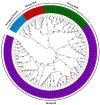
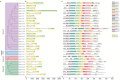
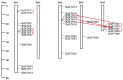
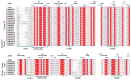
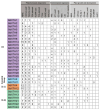
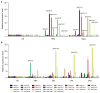
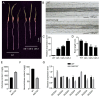
Xyloglucan endotransglucosylase/hydrolases (XTHs) are a class of cell wall-associated enzymes involved in the construction and remodeling of cellulose/xyloglucan crosslinks. However, knowledge of this gene family in the model monocot Brachypodium distachyon is limited. A total of 29 BdXTH genes were identified from the whole genome, and these were further divided into three subgroups (Group I/II, Group III, and the Ancestral Group) through evolutionary analysis. Gene structure and protein motif analyses indicate that closely clustered BdXTH genes are relatively conserved within each group. A highly conserved amino acid domain (DEIDFEFLG) responsible for catalytic activity was identified in all BdXTH proteins. We detected three pairs of segmentally duplicated BdXTH genes and five groups of tandemly duplicated BdXTH genes, which played vital roles in the expansion of the BdXTH gene family. Cis-elements related to hormones, growth, and abiotic stress responses were identified in the promoters of each BdXTH gene, and when roots were treated with two abiotic stresses (salinity and drought) and four plant hormones (IAA, auxin; GA3, gibberellin; ABA, abscisic acid; and BR, brassinolide), the expression levels of many BdXTH genes changed significantly. Transcriptional analyses of the BdXTH genes in 38 tissue samples from the publicly available RNA-seq data indicated that most BdXTH genes have distinct expression patterns in different tissues and at different growth stages. Overexpressing the BdXTH27 gene in Brachypodium led to reduced root length in transgenic plants, which exhibited higher cellulose levels but lower hemicellulose levels compared to wild-type plants. Our results provide valuable information for further elucidation of the biological functions of BdXTH genes in the model grass B. distachyon.
Keywords: Brachypodium distachyon; gene expression; genome-wide analysis; xyloglucan endotransglucosylase/hydrolase.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
-
- Yokoyama R., Nishitani K. A comprehensive expression analysis of all members of a gene family encoding cell-wall enzymes allowed us to predict cis-regulatory regions involved in cell-wall construction in specific organs of Arabidopsis. Plant Cell Physiol. 2001;42:1025–1033. doi: 10.1093/pcp/pce154. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

