GnomAD Missense Variants of Uncertain Significance: Implications for p53 Stability and Phosphorylation
- PMID: 40806592
- PMCID: PMC12347906
- DOI: 10.3390/ijms26157455
GnomAD Missense Variants of Uncertain Significance: Implications for p53 Stability and Phosphorylation
Abstract
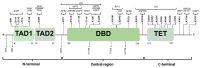
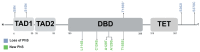
The TP53 gene, frequently mutated across multiple cancer types, plays a pivotal role in regulating the cell cycle and apoptosis through its protein, p53. Missense variants of uncertain significance (VUSs) in TP53 present challenges in understanding their impact on protein function and complicate clinical interpretation. This study aims to analyze the effects of missense VUSs in p53, as reported in the gnomAD database, with a specific focus on their impact on protein stability and phosphorylation. In this study, 33 missense VUSs in TP53 reported in the gnomAD database were analyzed using in silico tools, including PhosphositePlus v6.7.4, the Kinase Library v0.0.11, and Dynamut2. Of these analyzed variants, five disrupted known phosphorylation sites, while another five created new consensus sequences for phosphorylation. Moreover, 20 variants exhibited a moderate destabilizing effect on the protein structure. At least three missense VUSs were identified as potentially affecting p53 function, which may contribute to cancer development. These findings highlight the importance of integrating in silico structural and functional analysis to assess the pathogenic potential of missense VUSs.
Keywords: TP53 gene; VUS; gnomAD; phosphorylation sites; protein stability.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
[A child with Fructose-1,6-bisphosphatase deficiency due to variant of FBP1 gene: Genetic and clinical analysis and literature review].Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2025 Jun 10;42(6):719-728. doi: 10.3760/cma.j.cn511374-20241011-00529. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2025. PMID: 40763970 Review. Chinese.
-
Prioritizing disease-associated missense variants with chemoproteomic-detected amino acids.Am J Hum Genet. 2025 Jul 3;112(7):1649-1663. doi: 10.1016/j.ajhg.2025.04.017. Epub 2025 May 23. Am J Hum Genet. 2025. PMID: 40412387 Free PMC article.
-
Investigation of Structural Mechanisms Underlying p53 Dysfunction Caused by 148 Missense Mutations Using AlphaFold3 and Molecular Dynamics Simulations.J Chem Inf Model. 2025 Jun 23;65(12):6322-6330. doi: 10.1021/acs.jcim.5c00580. Epub 2025 Jun 4. J Chem Inf Model. 2025. PMID: 40464633
-
Falls prevention interventions for community-dwelling older adults: systematic review and meta-analysis of benefits, harms, and patient values and preferences.Syst Rev. 2024 Nov 26;13(1):289. doi: 10.1186/s13643-024-02681-3. Syst Rev. 2024. PMID: 39593159 Free PMC article.
References
-
- Chen S., Francioli L.C., Goodrich J.K., Collins R.L., Kanai M., Wang Q., Alföldi J., Watts N.A., Vittal C., Gauthier L.D., et al. A genomic mutational constraint map using variation in 76,156 human genomes. Nature. 2023;626:92–100. doi: 10.1038/s41563-022-01381-4. Erratum in Nature 2024, 625, 92–100. - DOI - PMC - PubMed
-
- Hu C., Susswein L.R., Roberts M.E., Yang H., Marshall M.L., Hiraki S., Berkofsky-Fessler W., Gupta S., Shen W., Dunn C.A., et al. Classification of BRCA2 Variants of Uncertain Significance (VUS) using an ACMG/AMP model incorporating a homology-directed repair (HDR) functional assay. Clin. Cancer Res. 2022;28:3742–3751. doi: 10.1158/1078-0432.CCR-22-0203. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

