Genome-Editing Tools for Lactic Acid Bacteria: Past Achievements, Current Platforms, and Future Directions
- PMID: 40806610
- PMCID: PMC12347395
- DOI: 10.3390/ijms26157483
Genome-Editing Tools for Lactic Acid Bacteria: Past Achievements, Current Platforms, and Future Directions
Abstract
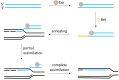
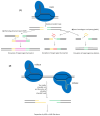
Lactic acid bacteria (LAB) are central to food, feed, and health biotechnology, yet their genomes have long resisted rapid, precise manipulation. This review charts the evolution of LAB genome-editing strategies from labor-intensive RecA-dependent double-crossovers to state-of-the-art CRISPR and CRISPR-associated transposase systems. Native homologous recombination, transposon mutagenesis, and phage-derived recombineering opened the door to targeted gene disruption, but low efficiencies and marker footprints limited throughput. Recent phage RecT/RecE-mediated recombineering and CRISPR/Cas counter-selection now enable scar-less point edits, seamless deletions, and multi-kilobase insertions at efficiencies approaching model organisms. Endogenous Cas9 systems, dCas-based CRISPR interference, and CRISPR-guided transposases further extend the toolbox, allowing multiplex knockouts, precise single-base mutations, conditional knockdowns, and payloads up to 10 kb. The remaining hurdles include strain-specific barriers, reliance on selection markers for large edits, and the limited host-range of recombinases. Nevertheless, convergence of phage enzymes, CRISPR counter-selection and high-throughput oligo recombineering is rapidly transforming LAB into versatile chassis for cell-factory and therapeutic applications.
Keywords: CRISPR-transposase; CRISPR/Cas9; Cre/lox; food biotechnology; genome editing; homologous recombination; lactic acid bacteria; probiotic engineering; recombineering; transposon mutagenesis.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

