Liquid Biopsy and Epigenetic Signatures in AML, ALL, and CNS Tumors: Diagnostic and Monitoring Perspectives
- PMID: 40806675
- PMCID: PMC12346961
- DOI: 10.3390/ijms26157547
Liquid Biopsy and Epigenetic Signatures in AML, ALL, and CNS Tumors: Diagnostic and Monitoring Perspectives
Abstract
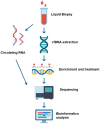
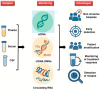
To deliver the most effective cancer treatment, clinicians require rapid and accurate diagnoses that delineate tumor type, stage, and prognosis. Consequently, minimizing the need for repetitive and invasive procedures like biopsies and myelograms, along with their associated risks, is a critical challenge. Non-invasive monitoring offers a promising avenue for tumor detection, screening, and prognostication. While the identification of oncogenes and biomarkers from circulating tumor cells or tissue biopsies is currently standard practice for cancer diagnosis and classification, accumulating evidence underscores the significant role of epigenetics in regulating stem cell fate, including proliferation, self-renewal, and malignant transformation. This highlights the importance of analyzing the methylome, exosomes, and circulating RNA for detecting cellular transformation. The development of diagnostic assays that integrate liquid biopsies with epigenetic analysis holds immense potential for revolutionizing tumor management by enabling rapid, non-invasive diagnosis, real-time monitoring, and personalized treatment decisions. This review covers current studies exploring the use of epigenetic regulation, specifically the methylome and circulating RNA, as diagnostic tools derived from liquid biopsies. This approach shows promise in facilitating the differentiation between primary central nervous system lymphoma and other central nervous system tumors and may enable the detection and monitoring of acute myeloid/lymphoid leukemia. We also discuss the current limitations hindering the rapid clinical translation of these technologies.
Keywords: PCNSL; circulating RNA; early diagnosis; epigenetic; leukemia; liquid biopsy; lymphoma; methylome.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
Can a Liquid Biopsy Detect Circulating Tumor DNA With Low-passage Whole-genome Sequencing in Patients With a Sarcoma? A Pilot Evaluation.Clin Orthop Relat Res. 2025 Jan 1;483(1):39-48. doi: 10.1097/CORR.0000000000003161. Epub 2024 Jun 21. Clin Orthop Relat Res. 2025. PMID: 38905450
-
Recent advances in liquid biopsy for precision oncology: emerging biomarkers and clinical applications in lung cancer.Future Oncol. 2025 Aug 5:1-19. doi: 10.1080/14796694.2025.2542051. Online ahead of print. Future Oncol. 2025. PMID: 40762271 Review.
-
Liquid biopsy - a narrative review with an update on current US governmental clinical trials targeting immunotherapy.Future Sci OA. 2025 Dec;11(1):2527598. doi: 10.1080/20565623.2025.2527598. Epub 2025 Aug 7. Future Sci OA. 2025. PMID: 40772765 Free PMC article. Review.
-
Management of urinary stones by experts in stone disease (ESD 2025).Arch Ital Urol Androl. 2025 Jun 30;97(2):14085. doi: 10.4081/aiua.2025.14085. Epub 2025 Jun 30. Arch Ital Urol Androl. 2025. PMID: 40583613 Review.
References
-
- Watanabe Y., Maekawa M. Advances in Clinical Chemistry. Volume 52. Elsevier; Amsterdam, The Netherlands: 2010. Methylation of DNA in Cancer; pp. 145–167. - PubMed
Publication types
LinkOut - more resources
Full Text Sources

