An Italian Single-Center Genomic Surveillance Study: Two-Year Analysis of SARS-CoV-2 Spike Protein Mutations
- PMID: 40806685
- PMCID: PMC12347092
- DOI: 10.3390/ijms26157558
An Italian Single-Center Genomic Surveillance Study: Two-Year Analysis of SARS-CoV-2 Spike Protein Mutations
Abstract
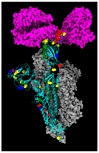
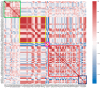
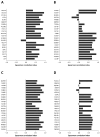
The repeated occurrence of SARS-CoV-2 variants, largely driven by virus-host interactions, was and will remain a public health concern. Spike protein mutations shaped viral infectivity, transmissibility, and immune escape. From February 2022 to April 2024, a local genomic surveillance program in Verona, Italy, was conducted on 1333 SARS-CoV-2-positive nasopharyngeal swabs via next generation full-length genome sequencing. Spike protein mutations were classified based on their prevalence over time. Mutations were grouped into five categories: fixed, emerging, fading, transient, and divergent. Notably, some divergent mutations displayed a "Lazarus effect," disappearing and later reappearing in new lineages, indicating potential adaptive advantages in specific genomic contexts. This two-year surveillance study highlights the dynamic nature of spike protein mutations and their role in SARS-CoV-2 evolution. The findings underscore the need for ongoing mutation-focused genomic monitoring to detect early signals of variant emergence, especially among mutations previously considered disadvantageous. Such efforts are critical for driving public health responses and guiding future vaccine and therapeutic strategies.
Keywords: SARS-CoV-2; genomic surveillance; mutational analysis; spike protein; whole genome sequencing.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
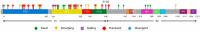




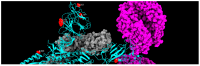
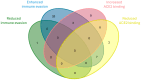
References
-
- Naqvi A.A.T., Fatima K., Mohammad T., Fatima U., Singh I.K., Singh A., Atif S.M., Hariprasad G., Hasan G.M., Hassan M.d.I. Insights into SARS-CoV-2 Genome, Structure, Evolution, Pathogenesis and Therapies: Structural Genomics Approach. Biochim. Et Biophys. Acta (BBA) Mol. Basis Dis. 2020;1866:165878. doi: 10.1016/j.bbadis.2020.165878. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- Department of Excellence 2023/2027, MUR, Italy/Ministero dell'università e della ricerca
- Quota FUR Prof. Gibellini/Department of Diagnostic and Public Health, Microbiology Section, University of Verona
- Quota FUR Virginia Lotti/Department of Diagnostic and Public Health, Microbiology Section, University of Verona
- Quota FUR Erica Diani/Department of Diagnostic and Public Health, Microbiology Section, University of Verona
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

