Genome-Wide Identification and Characterisation of the 4-Coumarate-CoA Ligase (4CL) Gene Family in Gastrodia elata and Their Transcriptional Response to Fungal Infection
- PMID: 40806741
- PMCID: PMC12347137
- DOI: 10.3390/ijms26157610
Genome-Wide Identification and Characterisation of the 4-Coumarate-CoA Ligase (4CL) Gene Family in Gastrodia elata and Their Transcriptional Response to Fungal Infection
Abstract
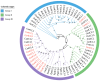
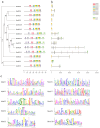
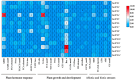
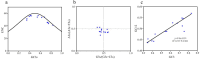
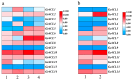
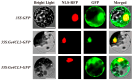
Gastrodia elata Blume is an important medicinal orchid, yet its large-scale cultivation is increasingly threatened by fungal diseases. The 4-coumarate-CoA ligase (4CL) gene family directs a key step in phenylpropanoid metabolism and plant defence, but its composition and function in G. elata have not been investigated. We mined the G. elata genome for 4CL homologues, mapped their chromosomal locations, and analysed their gene structures, conserved motifs, phylogenetic relationships, promoter cis-elements and codon usage bias. Publicly available transcriptomes were used to examine tissue-specific expression and responses to fungal infection. Subcellular localisation of selected proteins was verified by transient expression in Arabidopsis protoplasts. Fourteen Ge4CL genes were identified and grouped into three clades. Two members, Ge4CL2 and Ge4CL5, were strongly upregulated in tubers challenged with fungal pathogens. Ge4CL2 localised to the nucleus, whereas Ge4CL5 localised to both the nucleus and the cytoplasm. Codon usage analysis suggested that Escherichia coli and Oryza sativa are suitable heterologous hosts for Ge4CL expression. This study provides the first genome-wide catalogue of 4CL genes in G. elata and suggests that Ge4CL2 and Ge4CL5 may participate in antifungal defence, although functional confirmation is still required. The dataset furnishes a foundation for functional characterisation and the molecular breeding of disease-resistant G. elata cultivars.
Keywords: 4CL gene family; Gastrodia elata Blume; fungal infection; molecular characterisation; molecular defence; transcriptome analysis.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures


Similar articles
-
Comparative transcriptome analysis reveals the potential mechanism of seed germination promoted by trametenolic acid in Gastrodia elata Blume.Sci Rep. 2025 Jul 24;15(1):26869. doi: 10.1038/s41598-025-12269-z. Sci Rep. 2025. PMID: 40702084 Free PMC article.
-
Multi-Time Point Transcriptome Analysis and Functional Validation Revealed Bol4CL41 Negatively Regulates Black Rot Resistance in Cabbage.Int J Mol Sci. 2025 Jun 26;26(13):6179. doi: 10.3390/ijms26136179. Int J Mol Sci. 2025. PMID: 40649962 Free PMC article.
-
An enhanced gene function analysis platform for Gastrodia Elata integrating further omics data and analysis tools.BMC Plant Biol. 2025 Aug 22;25(1):1111. doi: 10.1186/s12870-025-07173-7. BMC Plant Biol. 2025. PMID: 40841935 Free PMC article.
-
Home treatment for mental health problems: a systematic review.Health Technol Assess. 2001;5(15):1-139. doi: 10.3310/hta5150. Health Technol Assess. 2001. PMID: 11532236
-
Immunogenicity and seroefficacy of pneumococcal conjugate vaccines: a systematic review and network meta-analysis.Health Technol Assess. 2024 Jul;28(34):1-109. doi: 10.3310/YWHA3079. Health Technol Assess. 2024. PMID: 39046101 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

