Multi-Component Synthesis of New Fluorinated-Pyrrolo[3,4- b]pyridin-5-ones Containing the 4-Amino-7-chloroquinoline Moiety and In Vitro-In Silico Studies Against Human SARS-CoV-2
- PMID: 40806777
- PMCID: PMC12347832
- DOI: 10.3390/ijms26157651
Multi-Component Synthesis of New Fluorinated-Pyrrolo[3,4- b]pyridin-5-ones Containing the 4-Amino-7-chloroquinoline Moiety and In Vitro-In Silico Studies Against Human SARS-CoV-2
Abstract
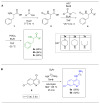
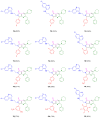
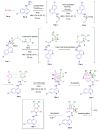
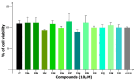
A one-pot synthetic methodology that combines an Ugi-Zhu three-component reaction (UZ-3CR) with a cascade sequence (intermolecular aza Diels-Alder cycloaddition/intramolecular N-acylation/decarboxylation/dehydration) using microwave-heating conditions, ytterbium (III) triflate (Yb(OTf)3) as the catalyst, and chlorobenzene (for the first time in a multi-component reaction (MCR)) as the solvent, was developed to synthesize twelve new fluorinated-pyrrolo[3,4-b]pyridin-5-ones containing a 4-amino-7-chloroquinoline moiety, yielding 50-77% in 95 min per product, with associated atom economies around 88%, also per product. Additionally, by in vitro tests, compounds 19d and 19i were found to effectively stop early SARS-CoV-2 replication, IC50 = 6.74 µM and 5.29 µM, at 0 h and 1 h respectively, while cell viability remained above 90% relative to the control vehicle at 10 µM. Additional computer-based studies revealed that the most active compounds formed strong favorable interactions with important viral proteins (Mpro, NTDα and NTDo) of coronavirus, supporting a two-pronged approach that affects both how the virus infects the cells and how it replicates its genetic material. Finally, quantum chemistry analyses of non-covalent interactions were performed from Density-Functional Theory (DFT) to better understand how the active compounds hit the virus.
Keywords: SARS-CoV-2; antiviral activity; drug design; fluorinated-compounds; multi-component reactions; polyheterocycles; pyrrolo[3,4-b]pyridin-5-ones; quinoline.
Conflict of interest statement
The author Nancy Viridiana Estrada-Toledo is employed by Health Pharma Professional Research S.A de C.V. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures



Similar articles
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
-
Synthesis of bis-furyl-pyrrolo[3,4-b]pyridin-5-ones via Ugi-Zhu reaction and in vitro activity assays against human SARS-CoV-2 and in silico studies on its main proteins.RSC Med Chem. 2022 Nov 18;14(1):154-165. doi: 10.1039/d2md00350c. eCollection 2023 Jan 25. RSC Med Chem. 2022. PMID: 36760742 Free PMC article.
-
Antiviral efficacy of hexane extract of Hypericum gaitii Haines against Chikungunya and SARS-CoV-2 viruses: in vitro and in silico approaches.J Ethnopharmacol. 2025 Jul 10;353(Pt A):120270. doi: 10.1016/j.jep.2025.120270. Online ahead of print. J Ethnopharmacol. 2025. PMID: 40651726
References
-
- Evans B.E., Rittle K.E., Bock M.G., DiPardo R.M., Freidinger R.M., Whiter W.L., Lundell G.F., Veber D.F., Anderson P.S., Chang R.S.L., et al. Methods for drug discovery: Development of potent, selective, orally effective cholecystokinin antagonists. J. Med. Chem. 1988;31:2235–2246. doi: 10.1021/jm00120a002. - DOI - PubMed
-
- Patchett A.A., Nargund R.P. Chapter 26. Privileged structures-An update. Annu. Rep. Med. Chem. 2000;35:289–298.
-
- Barreiro E.J. Privileged Scaffolds in Medicinal Chemistry: An introduction. In: Bräse S., editor. Privileged Scaffolds in Medicinal Chemistry: Design, Synthesis, Evaluation. Royal Society of Chemistry; Cambridge, UK: 2015. pp. 1–15.
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

