Incorporation and Repair of Epigenetic Intermediates as Potential Chemotherapy Agents
- PMID: 40807411
- PMCID: PMC12348197
- DOI: 10.3390/molecules30153239
Incorporation and Repair of Epigenetic Intermediates as Potential Chemotherapy Agents
Abstract
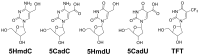
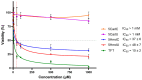
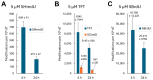
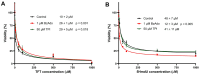
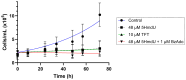
The incorporation of nucleoside analogs into DNA by polymerases, followed by their removal through base excision repair (BER), represents a promising strategy for cancer chemotherapy. In this study, we investigated the incorporation and cytotoxic effects of several nucleoside analogs-some of which are epigenetic reprogramming intermediates-in the U87 glioblastoma cell line. We found that two analogs, 5-hydroxymethyl-2'-deoxyuridine (5HmdU) and trifluorothymidine (TFT), are both cytotoxic and are efficiently incorporated into genomic DNA. In contrast, the 5-carboxy analogs-5-carboxy-2'-deoxyuridine (5CadU) and 5-carboxycytidine (5CadC)-showed no cytotoxicity and were not incorporated into DNA. Interestingly, 5-hydroxymethyl-2'-deoxycytidine (5HmdC) was cytotoxic but was not directly incorporated into DNA. Instead, it was deaminated into 5HmdU, which was then incorporated and likely responsible for the observed toxicity. 5HmdU is actively removed from DNA through the BER pathways. In contrast, TFT remains stably incorporated and is neither excised by BER nor does it hydrolyze into 5CadU-a known substrate for the DNA glycosylase SMUG1. We also found that N6-benzyladenosine (BzAdo), an inhibitor of the enzyme 2'-deoxynucleoside 5'-phosphate N-hydrolase (DNPH1), enhances the cytotoxicity of 5HmdU. However, the thymidine phosphorylase inhibitor tipiracil hydrochloride (TPI) does not increase the cytotoxic effect of TFT in U87 cells. Together, these findings highlight 5HmdU and TFT as promising chemotherapeutic agents for glioblastoma, each with distinct mechanisms of action and cellular processing.
Keywords: 5-hydroxymethyuracil; 5HmdU; BER; DNA; SMUG1; TFT; chemotherapy; epigenetic intermediates; glioblastoma; trifluorothymidine.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures



Similar articles
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
Short-Term Memory Impairment.2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 31424720 Free Books & Documents.
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
-
Systemic treatments for metastatic cutaneous melanoma.Cochrane Database Syst Rev. 2018 Feb 6;2(2):CD011123. doi: 10.1002/14651858.CD011123.pub2. Cochrane Database Syst Rev. 2018. PMID: 29405038 Free PMC article.
-
A rapid and systematic review of the clinical effectiveness and cost-effectiveness of paclitaxel, docetaxel, gemcitabine and vinorelbine in non-small-cell lung cancer.Health Technol Assess. 2001;5(32):1-195. doi: 10.3310/hta5320. Health Technol Assess. 2001. PMID: 12065068
References
-
- Li L.S., Morales J.C., Veigl M., Sedwick D., Greer S., Meyers M., Wagner M., Fishel R., Boothman D.A. DNA mismatch repair (MMR)-dependent 5-fluorouracil cytotoxicity and the potential for new therapeutic targets. Br. J. Pharmacol. 2009;158:679–692. doi: 10.1111/j.1476-5381.2009.00423.x. - DOI - PMC - PubMed
-
- Iveland T.S., Hagen L., de Sousa M.M.L., Liabakk N.B., Aas P.A., Sharma A., Kavli B., Slupphaug G. Cytotoxic mechanisms of pemetrexed and HDAC inhibition in non-small cell lung cancer cells involving ribonucleotides in DNA. Sci. Rep. 2025;15:2082. doi: 10.1038/s41598-025-86007-w. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

