Microbial Dynamics in a Musalais Wine Fermentation: A Metagenomic Study
- PMID: 40807507
- PMCID: PMC12346735
- DOI: 10.3390/foods14152570
Microbial Dynamics in a Musalais Wine Fermentation: A Metagenomic Study
Abstract
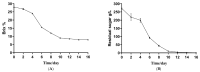
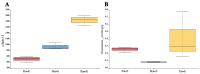
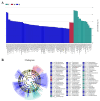
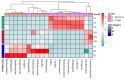
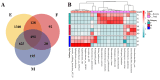
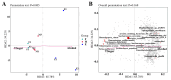
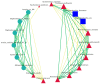
This study provides a comprehensive analysis of the microbial dynamics involved in the fermentation process of traditional Musalais wine, an intangible cultural heritage of Xinjiang. Utilizing metagenomic sequencing, we identified 2894 microbial species, of which 494 persisted throughout the fermentation process. Saccharomyces cerevisiae was the dominant species, with its prevalence increasing from 97.35% in the early phase to 99.38% in the mid phase, before slightly decreasing to 98.79% in the late phase. Additionally, 24 non-Saccharomyces yeast species, including Hanseniaspora uvarum, Lachancea thermotolerans, and Torulaspora delbrueckii, were detected. Common species associated with other fermented foods, including Wickerhamomyces anomalus, Kluyveromyces marxianus, Saccharomyces eubayanus, and Zygosaccharomyces parabailii, were also identified. Notably, species not previously used in food fermentation, such as Saccharomyces jurei, Sodiomyces alkalinus, Vanrija pseudolonga, and Moesziomyces antarcticus, were also identified in this study. Furthermore, the Kyoto Encyclopedia of Genes and Genomes (KO) and Gene Ontology (GO) revealed notable variations in metabolic pathways and enriched functional genes. In addition, a total of 82 volatile compounds were detected in the final product, with higher alcohols (60.12%), esters (37.80%), and organic acids (1.80%) being the most prevalent. These results offer important insights into microbial interactions and their influence on Musalais wine quality, laying the groundwork for optimizing the fermentation process.
Keywords: Musalais wine; metagenomics; microbial diversity; spontaneous/wild fermentation; volatile compounds.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
An integrated multiphase dynamic genome-scale model explains batch fermentations led by species of the Saccharomyces genus.mSystems. 2025 Feb 18;10(2):e0161524. doi: 10.1128/msystems.01615-24. Epub 2025 Jan 22. mSystems. 2025. PMID: 39840996 Free PMC article.
-
A Rapid Growth Rate Underpins the Dominance of Hanseniaspora uvarum in Spontaneous Grape Juice Fermentations.Yeast. 2025 Jun;42(5-7):116-125. doi: 10.1002/yea.4000. Epub 2025 Mar 21. Yeast. 2025. PMID: 40116327
-
Thriving in Adversity: Yeasts in the Agave Fermentation Environment.Yeast. 2025 Jan;42(1-3):16-30. doi: 10.1002/yea.3989. Epub 2025 Feb 19. Yeast. 2025. PMID: 39967574 Free PMC article. Review.
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
References
-
- Zhu L.X., Wang L.L., Song H.Z., Guo D.Q., Fan Y.G., Hou C.H., Xue J.L. Qualitative analysis of the main aroma compounds associated with traditional Musalais processing in Xinjiang, China. J. Inst. Brew. 2012;118:236–242. doi: 10.1002/jib.26. - DOI
-
- Ma W.R., Yu J.J., Yang F., Zhang X.M., Zhang F.J., Jin W.Y., Sun Z.W., Zhao Z.H., Jia S.R., Zhong C., et al. Metagenomic analysis of the relationship between the microorganisms and the volatiles’ development in the wines during spontaneous fermentation from the eastern foothills of the Ningxia Helan mountains in China. J. Sci. Food Agric. 2023;103:6429–6439. doi: 10.1002/jsfa.12718. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

