Occurrence of Aspergillus and Penicillium Species, Accumulation of Fungal Secondary Metabolites, and qPCR Detection of Potential Aflatoxigenic Aspergillus Species in Chickpea (Cicer arietinum L.) Seeds from Different Farming Systems
- PMID: 40807547
- PMCID: PMC12346705
- DOI: 10.3390/foods14152610
Occurrence of Aspergillus and Penicillium Species, Accumulation of Fungal Secondary Metabolites, and qPCR Detection of Potential Aflatoxigenic Aspergillus Species in Chickpea (Cicer arietinum L.) Seeds from Different Farming Systems
Abstract
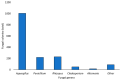
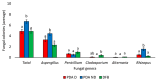
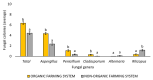
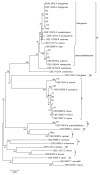
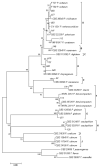
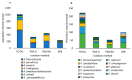
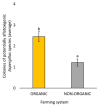
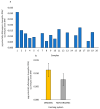
The European chickpea market raises concerns about health risks for consumers due to contamination by mycotoxins. Contamination levels can vary depending on the farming system, and rapid and reliable screening tools are desirable. In this study, marketed chickpea seed samples from organic and non-organic farming systems were analyzed for fungal and mycotoxin contamination. Aspergillus and Penicillium were the most frequently identified mycotoxigenic genera. Significant differences in fungal detection were observed among the three isolation methods used, whose combined application is proposed to enhance detection efficiency. The number of Aspergillus and Penicillium colonies was significantly higher in the organic samples. Molecular analysis identified different species within each genus, including several not previously reported in chickpea, as well as potentially aflatoxigenic species such as A. flavus/oryzae and A. parasiticus. LC-MS/MS analysis revealed aflatoxin production only by A. parasiticus, which was present in low amounts. However, the presence of potentially aflatoxigenic Aspergillus species suggests that chickpeas should be monitored to detect their safety and subsequently protect consumer health. A qPCR protocol targeting the omt-1 gene, involved in aflatoxin biosynthesis, proved to be a promising rapid tool for detecting potentially aflatoxigenic Aspergillus species.
Keywords: aflatoxins; mycobiota; mycotoxigenic fungi; mycotoxins; organic farming system; post-harvest; pulses.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
Aspergillus species and aflatoxin contamination of tobacco snuff samples sold in some parts of southeastern Nigeria.Mycotoxin Res. 2025 Aug;41(3):415-423. doi: 10.1007/s12550-025-00590-5. Epub 2025 May 7. Mycotoxin Res. 2025. PMID: 40332655
-
A rapid and systematic review of the clinical effectiveness and cost-effectiveness of paclitaxel, docetaxel, gemcitabine and vinorelbine in non-small-cell lung cancer.Health Technol Assess. 2001;5(32):1-195. doi: 10.3310/hta5320. Health Technol Assess. 2001. PMID: 12065068
-
Suppressed Production of Aflatoxin B1 by Aspergillus flavus and Aspergillus parasiticus on California In-hull Almonds and Hull Fragments.J Food Prot. 2025 Jun 23;88(7):100531. doi: 10.1016/j.jfp.2025.100531. Epub 2025 May 6. J Food Prot. 2025. PMID: 40339989
-
Management of urinary stones by experts in stone disease (ESD 2025).Arch Ital Urol Androl. 2025 Jun 30;97(2):14085. doi: 10.4081/aiua.2025.14085. Epub 2025 Jun 30. Arch Ital Urol Androl. 2025. PMID: 40583613 Review.
References
-
- FAOSTAT. 2025. [(accessed on 26 February 2025)]. Available online: https://www.fao.org/faostat/en/#data/QCL.
-
- Merga B., Haji J. Economic importance of chickpea: Production, value, and world trade. Cogent. Food Agric. 2019;5:1615718. doi: 10.1080/23311932.2019.1615718. - DOI
-
- Cid-Gallegos M.S., Sánchez-Chino X.M., Álvarez-González I., Madrigal-Bujaidar E., Vásques-Garzón V.R., Baltiérrez-Hoyos R., Villa-Treviño S., Dávila-Ortíz G., Jiménez-Martinéz C. Modification of in vitro and in vivo antioxidant activity by consumption of cooked chickpea in a colon cancer model. Nutrients. 2020;12:2572. doi: 10.3390/nu12092572. - DOI - PMC - PubMed
-
- Mahbub R., Francis N., Blanchard C., Santhakumar A. The anti-inflammatory and antioxidant properties of chickpea hull phenolic extracts. Food Biosci. 2021;40:100850. doi: 10.1016/j.fbio.2020.100850. - DOI
LinkOut - more resources
Full Text Sources

