The miR172a-ERF416/413 module regulates soybean seed traits
- PMID: 40808470
- PMCID: PMC12590345
- DOI: 10.1111/jipb.70015
The miR172a-ERF416/413 module regulates soybean seed traits
Abstract
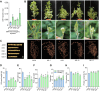
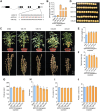
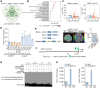
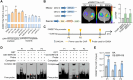
Soybean (Glycine max) provides vegetable oils and proteins for human consumption. Its production depends on seeds and other production-related agronomic traits. How the seed traits are regulated in soybean remains largely unclear. In this study, we identified a miR172a-ERF416/413 module for the regulation of seed traits. The miR172a can cleave the targets ERF416 and ERF413 to affect the downstream gene expression for the reduction of soybean seed size and weight. Both the MIR172a-overexpressing transgenic soybean plants and the erf416/413 mutants produced smaller seeds than the control. Consistently, the ERF416-overexpressing transgenic soybean plants generated larger seeds. ERF416 and ERF413 were directly targeted to the promoter of GmKIX8-1 and GmSWEET10a to regulate their gene expression for seed size/weight control. Interestingly, the erf416/413 mutants showed higher seed yield per plant and higher total seed fatty acid (FA) content, whereas the MIR172a-transgenic soybean had lower total seed FA content compared with the control cultivar, suggesting that miR172a and ERF416/413 may function in FA accumulation through different pathways. Haplotypes of the ERF416 promoter region were further analyzed and Hap1 was correlated with higher gene expression and higher seed weight, while Hap3 was correlated with higher total seed lipid content. Our study revealed a new module for seed trait control. Manipulation of such alleles should facilitate breeding for high-oil and high-yield soybean cultivars.
Keywords: ERF416/ERF413 transcription factors; fatty acids; miR172a; seed size/weight; soybean.
© 2025 The Author(s). Journal of Integrative Plant Biology published by John Wiley & Sons Australia, Ltd on behalf of Institute of Botany, Chinese Academy of Sciences.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Chen, X. , Liu, C. , Guo, P. , Hao, X. , Pan, Y. , Zhang, K. , Liu, W. , Zhao, L. , Luo, W. , He, J. , et al. (2023). Differential SW16.1 allelic effects and genetic backgrounds contributed to increased seed weight after soybean domestication. J. Integr. Plant Biol. 65: 1734–1752. - PubMed
MeSH terms
Substances
Grants and funding
- 2024ZD04078/Biological Breeding-National Science and Technology Major Project
- 32090062/National Natural Science Foundation of China
- 32090063/National Natural Science Foundation of China
- 32171930/National Natural Science Foundation of China
- 2021YFF1000104/National Key Research and Development Program of China
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

