Machine learning-based drug-drug interaction prediction: a critical review of models, limitations, and data challenges
- PMID: 40808680
- PMCID: PMC12344460
- DOI: 10.3389/fphar.2025.1632775
Machine learning-based drug-drug interaction prediction: a critical review of models, limitations, and data challenges
Abstract
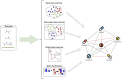
Background/objectives: New computational methods, based on statistical, machine learning, and deep learning techniques using drug-related entities (e.g., genes, protein bindings, etc.), help reduce the costs of in-vitro experiments through drug-drug interaction prediction (DDIp). This review examines recent advances in DDIp. It presents an in-depth review of the state-of-the-art studies relating to semi-supervised, supervised, self-supervised learning, and other techniques such as graph-based learning and matrix factorization methods for predicting DDIs. All possible interactions between drugs are not known, and accurately predicting interactions is even more difficult due to the complex nature of drug-drug interactions (DDI).
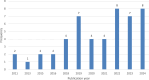
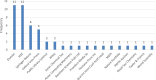
Methods: Of the 49 papers published in Web of Science in the last 6 years, 24 papers were considered relevant based on information presented in their titles and abstracts. The included articles focus specifically on predicting DDIs using a type of machine learning algorithm. Excluded articles focused on drug discovery, drug repurposing, molecular representation, or the extraction of biomedical interactions. The methodology, results limitations, and future research directions were studied for each paper. Common challenges, limitations, and future research directions were analyzed.
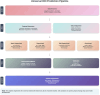
Results and conclusion: The main limitations are class imbalance, poor performance on new drugs, limited explainability, and the need for additional data sources.
Keywords: adverse drug reactions; drug-drug interaction; graph-based learning; healthcare; machine learning techniques; semi-supervised learning; supervised learning.
Copyright © 2025 Gheorghita, Bocanet and Iantovics.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
A rapid and systematic review of the clinical effectiveness and cost-effectiveness of topotecan for ovarian cancer.Health Technol Assess. 2001;5(28):1-110. doi: 10.3310/hta5280. Health Technol Assess. 2001. PMID: 11701100
-
Health professionals' experience of teamwork education in acute hospital settings: a systematic review of qualitative literature.JBI Database System Rev Implement Rep. 2016 Apr;14(4):96-137. doi: 10.11124/JBISRIR-2016-1843. JBI Database System Rev Implement Rep. 2016. PMID: 27532314
-
Approaches for predicting dairy cattle methane emissions: from traditional methods to machine learning.J Anim Sci. 2024 Jan 3;102:skae219. doi: 10.1093/jas/skae219. J Anim Sci. 2024. PMID: 39123286 Free PMC article.
-
[Volume and health outcomes: evidence from systematic reviews and from evaluation of Italian hospital data].Epidemiol Prev. 2013 Mar-Jun;37(2-3 Suppl 2):1-100. Epidemiol Prev. 2013. PMID: 23851286 Italian.
References
-
- Cao J., Lin X., Guo S., Liu L., Liu T., Wang B. (2021). “Bipartite graph embedding via mutual information maximization,” in Proceedings of the 14th ACM international conference on web search and data mining (New York, NY, USA: Association for Computing Machinery; ), 635–643. 10.1145/3437963.3441783 - DOI
-
- Chandra Umakantham O., Srinivasan S., Pathak V. (2024). Detecting side effects of adverse drug reactions through drug-drug interactions using graph neural networks and self-supervised learning. IEEE Access 12, 93823–93840. 10.1109/ACCESS.2024.3407877 - DOI
-
- Chen T., Guestrin C. (2016). “XGBoost: a scalable tree boosting system,” in Proceedings of the 22nd ACM SIGKDD international conference on knowledge discovery and data mining (New York, NY, USA: Association for Computing Machinery; ), 785–794. 10.1145/2939672.2939785 - DOI
-
- Chu X., Lin Y., Wang Y., Wang L., Wang J., Gao J. (2019). MLRDA: a multi-task semi-supervised learning framework for drug-drug interaction prediction, 4518–4524.
Publication types
LinkOut - more resources
Full Text Sources

