Experimental validation of cuproptosis-associated molecular signatures and their immunological implications in pulmonary tuberculosis
- PMID: 40808944
- PMCID: PMC12343668
- DOI: 10.3389/fimmu.2025.1570992
Experimental validation of cuproptosis-associated molecular signatures and their immunological implications in pulmonary tuberculosis
Abstract
Background: The pathogenic mechanism underlying Mycobacterium tuberculosis (MTB) remains elusive, posing challenges to its diagnosis and treatment. Cuproptosis is a newly identified mechanism of cell death. This study explores the role of cuproptosis-related genes (CRGs) in pulmonary tuberculosis (PTB) to uncover potential diagnostic biomarkers and therapeutic targets.
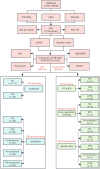
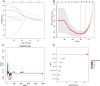
Methods: Differentially expressed gene (DEG) analysis and weighted gene co-expression network analysis (WGCNA) were carried out using the GSE83456 dataset. PTB-associated DEGs were intersected with CRGs to identify PTB-related CRGs. Subsequent analyses included functional enrichment, gene interaction, and protein-protein interaction (PPI) network construction. Hub CRGs were screened out via least absolute shrinkage and selection operator (LASSO) regression and random forest (RF) algorithms. Diagnostic models were subsequently constructed and validated. The associations of immune cell infiltration and pathway with the identified hub genes were evaluated through single-sample gene set enrichment analysis (ssGSEA) and CIBERSORT. Hub gene expressions were validated in the GSE42834 and GSE89403 datasets, as well as by RT-qPCR and Western blot (WB) in PTB and extrapulmonary tuberculosis (EPTB) patients. The GSE89403 dataset and gene expression profiling were leveraged to analyze the differential expression of hub genes and their dynamic changes during treatment.
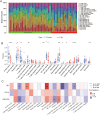
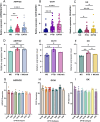
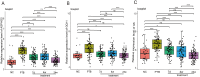
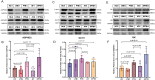
Results: Seven PTB-related CRGs were significantly upregulated, were significantly upregulated, among which ASPHD2, GK, and GCH1 were identified as hub genes. These genes exhibited high expression levels in patients with PTB and EPTB, with marked reductions observed following treatment. Notable alterations in immune cell infiltration and immune function in PTB patients were closely related to these hub genes, suggesting activation of innate immune responses and suppression of adaptive immune function.
Conclusion: The cuproptosis hub genes ASPHD2, GK, and GCH1 influence the pathogenesis of PTB, and possibly serve as novel diagnostic biomarkers and therapeutic targets.
Keywords: cuproptosis; experimental validation; gene expression analysis; immune dysregulation; pulmonary tuberculosis.
Copyright © 2025 Liu, Ma, Li, Xue, Mi, Li, Bai, Guo, Liu, Liang, Liang and Wu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






References
-
- WHO . Global tuberculosis report 2024. Geneva: World Health Organization; (2024).
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

