Combining Phenotypic and Genotypic Methods for the Rapid Detection of Antibiotic-Resistant Bacteria in Wastewater Using a Microfluidic Centrifugal Disc
- PMID: 40809007
- PMCID: PMC12340755
- DOI: 10.1021/acsestengg.5c00107
Combining Phenotypic and Genotypic Methods for the Rapid Detection of Antibiotic-Resistant Bacteria in Wastewater Using a Microfluidic Centrifugal Disc
Abstract
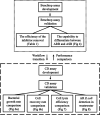
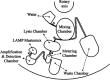
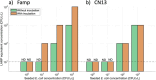
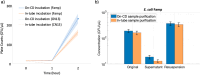
Antimicrobial resistance (AMR) has emerged as a critical global health threat, leading to untreatable infectious diseases and significantly increasing human morbidity and mortality. Rapid, portable, and user-friendly detection of antibiotic-resistant bacteria (ARB) in the environment plays a key role in controlling the spread of diseases and combating the emergence of AMR. In this study, we report a portable microfluidic centrifugal disc (CD) assay incorporating both phenotypic and genotypic methods for ARB detection. This assay significantly reduces the time required for bacterial culturing from days to hours while maintaining the reliability of phenotypic methods. Moreover, the CD assay eliminates the need for a centralized laboratory and well-trained personnel. The on-CD ARB detection can be completed in less than 3 h after sample and reagent loading. The results demonstrated that the on-CD assay provided performance comparable to that of the benchtop ARB detection assay. Analysis of variance (ANOVA) comparing the benchtop and on-CD assays yielded a p-value of 0.08 for the bacterial growth rate and a p-value of 0.73 for the cell recovery rate after the sample purification, confirming no significant differences between the two approaches. The ARB detection assay was able to identify ampicillin-resistant spiked in wastewater at concentrations as low as 10 CFU/μL. Additionally, the CD assay detected indigenous ampicillin-resistant in wastewater, demonstrating the potential of this platform for environmental applications.
Keywords: ARB; ampicillin-resistant E. coli; microfluidic centrifugal disc; sample-to-answer.
© 2025 The Authors. Published by American Chemical Society.
Figures



Similar articles
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
Antimicrobial resistance in southeast Asian water environments: A systematic review of current evidence and future research directions.Sci Total Environ. 2023 Oct 20;896:165229. doi: 10.1016/j.scitotenv.2023.165229. Epub 2023 Jun 30. Sci Total Environ. 2023. PMID: 37394072
-
Xpert® MTB/RIF assay for pulmonary tuberculosis and rifampicin resistance in adults.Cochrane Database Syst Rev. 2014 Jan 21;2014(1):CD009593. doi: 10.1002/14651858.CD009593.pub3. Cochrane Database Syst Rev. 2014. Update in: Cochrane Database Syst Rev. 2019 Jun 07;6:CD009593. doi: 10.1002/14651858.CD009593.pub4. PMID: 24448973 Free PMC article. Updated.
-
Xpert® MTB/RIF assay for pulmonary tuberculosis and rifampicin resistance in adults.Cochrane Database Syst Rev. 2013 Jan 31;(1):CD009593. doi: 10.1002/14651858.CD009593.pub2. Cochrane Database Syst Rev. 2013. Update in: Cochrane Database Syst Rev. 2014 Jan 21;(1):CD009593. doi: 10.1002/14651858.CD009593.pub3. PMID: 23440842 Free PMC article. Updated.
-
Effect of ozonation-based disinfection methods on the removal of antibiotic resistant bacteria and resistance genes (ARB/ARGs) in water and wastewater treatment: a systematic review.Sci Total Environ. 2022 Mar 10;811:151404. doi: 10.1016/j.scitotenv.2021.151404. Epub 2021 Nov 9. Sci Total Environ. 2022. PMID: 34767893
References
-
- World Health Organization: WHO . Antimicrobial resistance. https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance. (accessed 01 29, 2025).
-
- Centers for Disease Control and Prevention (U.S.) . 2019 Antibiotic Resistance Threats Report; CDC, 2019. https://www.cdc.gov/antimicrobial-resistance/data-research/threats/index... (accessed 10 03 2025).
-
- The Review on Antimicrobial Resistance, Chaired by Jim O’Neill , 2014. Antimicrobial resistance: tackling a crisis for the health and wealth of nations. https://amr-review.org/sites/default/files/AMR%20Review%20Paper%20-%20Ta.... (accessed 01 29, 2025).
-
- Singh A. K., Kaur R., Verma S., Singh S.. Antimicrobials and Antibiotic Resistance Genes in Water Bodies: Pollution, Risk, and Control. Front. Environ. Sci. 2022;10:830861. doi: 10.3389/fenvs.2022.830861. - DOI
LinkOut - more resources
Full Text Sources
