Comparative genomic analysis reveals the adaptive traits of Ralstonia spp. in aquatic environments
- PMID: 40809044
- PMCID: PMC12345375
- DOI: 10.3389/fmicb.2025.1625651
Comparative genomic analysis reveals the adaptive traits of Ralstonia spp. in aquatic environments
Abstract
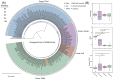
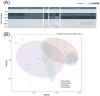
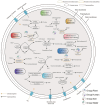
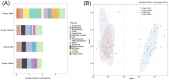
Ralstonia spp. are highly adaptable bacteria that are widely distributed across diverse environments. Here, we isolated four Ralstonia pickettii (R. pickettii) genomes from cultures of Dolichospermum spp., and using a comparative genomic framework of 228 Ralstonia genomes. We performed phylogenetic analyses that grouped them into water, soil, plant, and human-associated clades based on their predominant isolation habitats. Fluorescence in situ hybridization revealed minimal physical interactions between R. pickettii and cyanobacterial cells, indicating a commensal or independent ecological relationship. Distinct differences in carbohydrate-active enzymes (CAZymes) and secondary metabolite profiles were observed between water and human-associated dominant groups compared to plant-associated dominant groups, highlighting potential niche-specific adaptations. The water-associated dominant groups harbored antibiotic resistance genes, including CeoB and OXA-type β-lactamase genes. These genes are typically linked to human-associated strains, suggesting potential horizontal gene transfer or shared selective pressures, and the gene content of T3SS is reduced. Notably, water-associated dominant groups exhibited a unique pyrimidine degradation pathway, potentially enabling the utilization of exogenous pyrimidines to support survival in nutrient-limited aquatic environments. We propose that the gene content loss of T3SS and the acquisition of specialized metabolic pathways reflect adaptive strategies of Ralstonia spp. for thriving in aquatic free-living niches.
Keywords: Ralstonia; antibiotic resistance; comparative genomics; microbial evolution; pathogenic microorganisms.
Copyright © 2025 Liu, Mao, Li, Huo and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures


References
-
- Berg G. M., Jørgensen N. O. (2006). Purine and pyrimidine metabolism by estuarine bacteria. Aquat. Microb. Ecol. 42, 215–226. doi: 10.3354/ame042215, PMID: - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

