Multiple-omics analysis of three novel haloalkaliphilic species of Kocuria revealed that the phenolic acid-degrading abilities are ubiquitous in the genus
- PMID: 40809046
- PMCID: PMC12343579
- DOI: 10.3389/fmicb.2025.1626161
Multiple-omics analysis of three novel haloalkaliphilic species of Kocuria revealed that the phenolic acid-degrading abilities are ubiquitous in the genus
Abstract
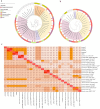
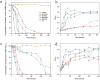
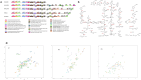
Phenolic acids (PAs), which can exert toxic effects on seed germination and plant growth, are the most common allelopathic substances found in soils. To better understand the degradation fates of PAs in the rhizosphere of halophytes, five haloalkaliphilic PA-degrading bacteria, which were identified as three novel species of Kocuria (namely, Kocuria rhizosphaerae sp. nov., Kocuria kalidii sp. nov., and Kocuria rhizosphaericola sp. nov.), were obtained from the rhizosphere and bulk soil of the halophyte Kalidium cuspidatum. All five Kocuria strains could efficiently degrade ferulic acid (FA) and cinnamic acid (CA) under saline-alkaline conditions. Genomic and transcriptomic analyses revealed that the acrylic groups of FA and CA were first converted to a carboxyl via the coenzyme A (CoA)-dependent non-β-oxidation pathway by the five Kocuria strains. However, the five Kocuria strains selected different aromatic ring-cleavage ways for the degradation of the benzoic derivatives intermediates of the two compounds. The protocatechuate result from FA was then thoroughly degraded through an aromatic ring-opening reaction catalyzed by protocatechuate 3,4-dioxygenase (PcaGH), and the β-ketoadipic acid pathway. At the same time, the yield of benzoate originated from CA was subsequently converted to catechol by the benzoate 1,2-dioxygenase system (BenABCD) or phenylacetyl-CoA epoxidase (PaaABCD) and further completed the ring-cleavage by catechol 1,2-dioxygenase or catechol 2,3-dioxygenase (two non-PcaGH dioxygenases). The comparative genomic analysis revealed that the genes for phenolic acids hydroxylation, protocatechuate 3,4-dioxygenation, and those involved in the β-ketoadipic acid pathways are universal in the Kocuria strains. It is also demonstrated that the Kocuria strains maintain their osmotic balance by accumulating potassium, rather than biosynthesizing organic osmoprotectants, under hypersaline conditions.
Keywords: Kocuria; biodegradation; comparative genomic analysis; osmotic stress; phenolic acids; polyphasic taxonomy.
Copyright © 2025 Xu, Sun, Zeng, Wei, Shen and Sun.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Sexual Harassment and Prevention Training.2024 Mar 29. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Mar 29. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 36508513 Free Books & Documents.
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
-
Abrocitinib, tralokinumab and upadacitinib for treating moderate-to-severe atopic dermatitis.Health Technol Assess. 2024 Jan;28(4):1-113. doi: 10.3310/LEXB9006. Health Technol Assess. 2024. PMID: 38343072 Free PMC article.
-
A rapid and systematic review of the clinical effectiveness and cost-effectiveness of paclitaxel, docetaxel, gemcitabine and vinorelbine in non-small-cell lung cancer.Health Technol Assess. 2001;5(32):1-195. doi: 10.3310/hta5320. Health Technol Assess. 2001. PMID: 12065068
-
Comparison of Two Modern Survival Prediction Tools, SORG-MLA and METSSS, in Patients With Symptomatic Long-bone Metastases Who Underwent Local Treatment With Surgery Followed by Radiotherapy and With Radiotherapy Alone.Clin Orthop Relat Res. 2024 Dec 1;482(12):2193-2208. doi: 10.1097/CORR.0000000000003185. Epub 2024 Jul 23. Clin Orthop Relat Res. 2024. PMID: 39051924
References
-
- Barillot C. D. C., Sarde C. -O., Bert V., Tarnaud E., Cochet N. (2013). A standardized method for the sampling of rhizosphere and rhizoplan soil bacteria associated to a herbaceous root system. Ann. Microbiol. 63, 471–476. doi: 10.1007/s13213-012-0491-y - DOI
