Biotype and host relatedness influence the composition of bacterial microbiomes in Schizaphis graminum aphids
- PMID: 40809050
- PMCID: PMC12345607
- DOI: 10.3389/fmicb.2025.1614492
Biotype and host relatedness influence the composition of bacterial microbiomes in Schizaphis graminum aphids
Abstract
Introduction: The microbiome of greenbug aphid (Schizaphis graminum (Rondani)) was investigated in regard to greenbug biotype, collection date, host species, and host cultivar.
Methods: DNA samples were collected from biotypes E and K feeding on 17 cultivars belonging to five host plant species, namely wheat, barley, rye, sorghum, and the goatgrass Aegilops triuncialis. Samples were taken immediately before infestation and two, four, and eight days thereafter. The V5-V7 hypervariable region of 16S rDNA was PCR amplified, Illumina sequenced, and aligned to a curated database of bacterial 16S rDNA sequences.
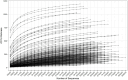
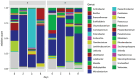
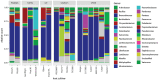
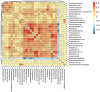
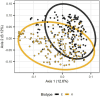
Results and discussion: The almost universal intracellular endosymbiont of aphids, Buchnera aphidicola, comprised 78.24 to 99.99% of the read counts among samples, largely because of its high copy number of genomes per bacteroid. Abundant non-Buchnera genera included Pseudomonas, Rhodanobacter, Massilia, and Enterobacter. Read counts of eight of 78 examined genera were more than 90% restricted to a single replicate of a single treatment. Shannon entropy was highest in biotype K and on the barley host, but it did not vary significantly among dates post infestation. Unweighted UniFrac distances most significantly varied with biotype, host plant species, infestation time, and almost all of their interactions. Weighted UniFrac and Jaccard distances varied less significantly. By counts of differentially populated genera, the factors biotype, host plant species, infestation time, and host plant resistance genes to greenbug, were consecutively less important. Functional analysis with PICRUSt2 illustrated a diminution of respiratory electron transport and long-chain fatty acids in the Buchnera endosymbiont, reflecting adaptation to an intracellular environment.
Keywords: 16S rRNA; RNA-seq; Schizaphis graminum; gut microflora; microbiome; symbiont.
Copyright This work is authored by Yan M. Crane, Charles F. Crane, Christian Webb and Brandon J. Schemerhorn on behalf of the U.S. Government and as regards Dr. Crane, Dr. Crane, Mr. Webb, Dr. Schemerhorn and the U.S. Government, is not subject to copyright protection in the United States. Foreign and other copyrights may apply.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Shotgun Metagenome Analysis of Two Schizaphis graminum Biotypes over Time With and Without Carried Cereal Yellow Dwarf Virus.Insects. 2025 May 23;16(6):554. doi: 10.3390/insects16060554. Insects. 2025. PMID: 40558984 Free PMC article.
-
Effects of host plants on aphid feeding behavior, fitness, and Buchnera aphidicola titer.Insect Sci. 2025 Jun;32(3):927-942. doi: 10.1111/1744-7917.13428. Epub 2024 Aug 8. Insect Sci. 2025. PMID: 39114883
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
Sertindole for schizophrenia.Cochrane Database Syst Rev. 2005 Jul 20;2005(3):CD001715. doi: 10.1002/14651858.CD001715.pub2. Cochrane Database Syst Rev. 2005. PMID: 16034864 Free PMC article.
-
A rapid and systematic review of the clinical effectiveness and cost-effectiveness of paclitaxel, docetaxel, gemcitabine and vinorelbine in non-small-cell lung cancer.Health Technol Assess. 2001;5(32):1-195. doi: 10.3310/hta5320. Health Technol Assess. 2001. PMID: 12065068
References
-
- Anderson M. J. (2001). A new method for non-parametric multivariate analysis of variance. Austral Ecol. 26, 32–46. doi: 10.1111/j.1442-9993.2001.01070.pp.x, PMID: - DOI
-
- Bowling R., Wilde G., Margolies D. (1998). Relative fitness of greenbug (Homoptera: Aphididae) biotypes E and I on sorghum, wheat, rye and barley. J. Econ. Entomol. 91, 1219–1223. doi: 10.1093/jee/91.5.1219 - DOI
LinkOut - more resources
Full Text Sources

