Distinct bacterial community structures and arsenic biotransformation gene profiles in dust
- PMID: 40809051
- PMCID: PMC12343739
- DOI: 10.3389/fmicb.2025.1607082
Distinct bacterial community structures and arsenic biotransformation gene profiles in dust
Abstract
Introduction: Microorganisms, which are ubiquitous in the environment, have evolved a diverse array of arsenic biotransformation genes (ABGs). Dust harbors a wide range of microorganisms. However, the distinct characteristics of bacterial community structures and ABG profiles in dust, compared with those in other environments such as soil and water, remain poorly understood.
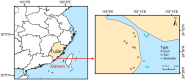
Methods: In this study, dust samples were simultaneously collected alongside surrounding soil and seawater samples in Xiamen, a coastal city of China, to investigate the distinct profiles and potential sources of bacterial communities and ABGs in dust using 16S rRNA gene amplicon sequencing and metagenomic sequencing.
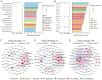
Results and discussion: Abundant and diverse bacterial communities and ABGs were detected in dust, revealing significant differences in community structures and ABG profiles compared with those in soil and seawater. Soil was identified as the primary source for both bacterial communities and ABGs in dust through fast expectation-maximization microbial source tracking (FEAST). Acetobacteraceae, which showed significantly greater relative abundance (p < 0.001) in dust than in soil and seawater, was also identified as a keystone taxon in the dust bacterial co-occurrence network. Furthermore, metagenome-assembled genomes (MAGs) affiliated with Acetobacteraceae were effectively recovered from dust via metagenomic binning, and these MAGs harbored an array of ABGs, indicating that Acetobacteraceae could be important hosts for ABGs in dust. Overall, our findings offer new insights into bacterial communities and ABGs in dust, thereby improving our understanding of arsenic biogeochemical cycling.
Keywords: arsenic; arsenic biotransformation genes; bacterial communities; dust; metagenomes.
Copyright © 2025 Yin, Lin, Hu, Yu, Sun and Yan.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
Systemic treatments for metastatic cutaneous melanoma.Cochrane Database Syst Rev. 2018 Feb 6;2(2):CD011123. doi: 10.1002/14651858.CD011123.pub2. Cochrane Database Syst Rev. 2018. PMID: 29405038 Free PMC article.
-
Sexual Harassment and Prevention Training.2024 Mar 29. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Mar 29. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 36508513 Free Books & Documents.
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
References
-
- Balakrishna G., Pervez S., Bisht D. (2011). Source apportionment of arsenic in atmospheric dust fall out in an urban residential area, Raipur, Central India. Atmos. Chem. Phys. 11, 5141–5151. doi: 10.5194/acp-11-5141-2011 - DOI
LinkOut - more resources
Full Text Sources

