Transcutaneous auricular vagus nerve stimulation regulates gut microbiota mediated peripheral inflammation and metabolic disorders to suppress depressive-like behaviors in CUMS rats
- PMID: 40809052
- PMCID: PMC12344421
- DOI: 10.3389/fmicb.2025.1576686
Transcutaneous auricular vagus nerve stimulation regulates gut microbiota mediated peripheral inflammation and metabolic disorders to suppress depressive-like behaviors in CUMS rats
Abstract
Background: Depression is a common mental disorder, and the changes of intestinal microflora and peripheral plasma metabolites can affect the gut-brain axis through vagus nerve, leading to the occurrence, and progress of the disease. Transcutaneous auricular vagus nerve stimulation (taVNS) has been previously shown to be clinically safe and effective in treating depression. However, there is no evidence whether its antidepressant effect is related to the regulation of intestinal flora and metabolites.
Objective: This study investigated the gut microbiota and plasma metabolism mechanisms of taVNS in the treatment of depression.
Methods: In this study, we established a chronic unpredictable mild stress (CUMS) model in SD rats for 5 weeks. During the last 3 weeks of CUMS treatment, the rats received continuous taVNS intervention for 3 weeks. Depressive-like behavior in SD rats was evaluated through behavioral assessments. The gut microbiota and plasma were analyzed using 16S rRNA gene sequencing and liquid chromatography-mass spectrometry (LC-MS) techniques.
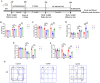
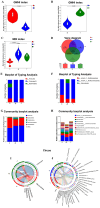
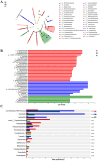
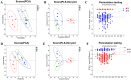
Results: Behavioral tests showed that taVNS significantly reversed the depressive-like behavior induced by CUMS in rats. 16S rRNA sequencing results showed that taVNS could improve the intestinal flora structure of CUMS rats. Microbial community characterization index showed that taVNS could reverse the gut microbiota dysbiosis in CUMS rats. ROC analysis revealed that Lachnospiraceae_NK4A136_group. Parabacteroides and Corynebacterium_1 are potential biomarkers for diagnosing gut microbiota dysbiosis in CUMS rats and could also serve as potential therapeutic targets for taVNS. Plasma metabolomics results showed that the differential metabolites between the CUMS group and the control group were primarily enriched in pathways such as bile acid metabolism, arachidonic acid metabolism and ether lipid metabolism. The differential metabolites between the taVNS group and the CUMS group were primarily enriched in pathways related to vitamin digestion and absorption, glycerophospholipid metabolism and amino acid metabolism. Correlation analysis between the gut microbiota and plasma metabolites suggested that pathogenic microbial genera such as Lachnospiraceae, Lactobacillus, and Tyzzerella were positively correlated with plasma metabolites during inflammation, bile acid, and lipid metabolism dysregulation, while beneficial microbiota showed the opposite trend.
Conclusion: This study demonstrated that taVNS can regulate the gut microbiota, including Lachnospiraceae, Lactobacillus, Tyzzerella, and Bacteroides genera, which mediate peripheral inflammation, bile acid, and lipid metabolism dysregulation, thereby reversing the depressive-like behavior induced by CUMS in rats and exerting an antidepressant effect.
Keywords: brain-gut axis; depression; gut microbiota; metabolomics; transcutaneous auricular vagus nerve stimulation.
Copyright © 2025 Zhang, He, Zhao, Zou, Liang, Zhang, Feng, Rong and Wang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures







Similar articles
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
Brainstem neuronal responses to transcutaneous auricular and cervical vagus nerve stimulation in rats.J Physiol. 2024 Aug;602(16):4027-4052. doi: 10.1113/JP286680. Epub 2024 Jul 19. J Physiol. 2024. PMID: 39031516 Free PMC article.
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
-
Quercetin alleviates chronic unpredictable mild stress-induced depression-like behavior by inhibiting NMDAR1 with α2δ-1 in rats.CNS Neurosci Ther. 2024 Apr;30(4):e14724. doi: 10.1111/cns.14724. CNS Neurosci Ther. 2024. PMID: 38615365 Free PMC article.
-
Transcutaneous auricular vagus nerve stimulation for epilepsy.Seizure. 2024 Jul;119:84-91. doi: 10.1016/j.seizure.2024.05.005. Epub 2024 May 30. Seizure. 2024. PMID: 38820674
References
LinkOut - more resources
Full Text Sources

