Transporter annotations are holding up progress in metabolic modeling
- PMID: 40809134
- PMCID: PMC12342017
- DOI: 10.3389/fsysb.2024.1394084
Transporter annotations are holding up progress in metabolic modeling
Abstract
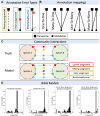
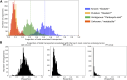
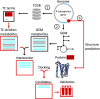
Mechanistic, constraint-based models of microbial isolates or communities are a staple in the metabolic analysis toolbox, but predictions about microbe-microbe and microbe-environment interactions are only as good as the accuracy of transporter annotations. A number of hurdles stand in the way of comprehensive functional assignments for membrane transporters. These include general or non-specific substrate assignments, ambiguity in the localization, directionality and reversibility of a transporter, and the many-to-many mapping of substrates, transporters and genes. In this perspective, we summarize progress in both experimental and computational approaches used to determine the function of transporters and consider paths forward that integrate both. Investment in accurate, high-throughput functional characterization is needed to train the next-generation of predictive tools toward genome-scale metabolic network reconstructions that better predict phenotypes and interactions. More reliable predictions in this domain will benefit fields ranging from personalized medicine to metabolic engineering to microbial ecology.
Keywords: flux balance analysis; functional genomics; metabolic modeling; microbial community modeling; transporter annotation.
Copyright © 2024 Casey, Bennion, D’haeseleer, Kimbrel, Marschmann and Navid.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
Short-Term Memory Impairment.2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 31424720 Free Books & Documents.
-
Fabricating mice and dementia: opening up relations in multi-species research.In: Jenkins N, Jack-Waugh A, Ritchie L, editors. Multi-Species Dementia Studies. Bristol (UK): Bristol University Press; 2025 Feb 25. Chapter 2. In: Jenkins N, Jack-Waugh A, Ritchie L, editors. Multi-Species Dementia Studies. Bristol (UK): Bristol University Press; 2025 Feb 25. Chapter 2. PMID: 40690569 Free Books & Documents. Review.
-
A rapid and systematic review of the clinical effectiveness and cost-effectiveness of paclitaxel, docetaxel, gemcitabine and vinorelbine in non-small-cell lung cancer.Health Technol Assess. 2001;5(32):1-195. doi: 10.3310/hta5320. Health Technol Assess. 2001. PMID: 12065068
-
The quantity, quality and findings of network meta-analyses evaluating the effectiveness of GLP-1 RAs for weight loss: a scoping review.Health Technol Assess. 2025 Jun 25:1-73. doi: 10.3310/SKHT8119. Online ahead of print. Health Technol Assess. 2025. PMID: 40580049 Free PMC article.
Cited by
-
Integrative genomic reconstruction reveals heterogeneity in carbohydrate utilization across human gut bifidobacteria.bioRxiv [Preprint]. 2025 May 17:2024.07.06.602360. doi: 10.1101/2024.07.06.602360. bioRxiv. 2025. Update in: Nat Microbiol. 2025 Aug;10(8):2031-2047. doi: 10.1038/s41564-025-02056-x. PMID: 39005317 Free PMC article. Updated. Preprint.
-
Recent trends in microbial production of alkanes.World J Microbiol Biotechnol. 2025 Sep 2;41(9):320. doi: 10.1007/s11274-025-04536-y. World J Microbiol Biotechnol. 2025. PMID: 40892357 Free PMC article. Review.
References
-
- Aplop F., Butler G. (2017). TransATH: transporter prediction via annotation transfer by homology. ARPN J. Eng. Appl. Sci. 12, 317–324.
LinkOut - more resources
Full Text Sources

