Diagnostic performance of metagenomics sequencing for pulmonary fungal infections: a clinical evaluation using the Nanopore platform
- PMID: 40809220
- PMCID: PMC12340274
- DOI: 10.21037/jtd-2025-1163
Diagnostic performance of metagenomics sequencing for pulmonary fungal infections: a clinical evaluation using the Nanopore platform
Abstract
Background: Pulmonary fungal infections are becoming increasingly prevalent, particularly among immunocompromised patients. Traditional culture-based and serological diagnostic methods exhibit low sensitivity and prolonged turnaround times (TATs), highlighting the need for more efficient diagnostic approaches. This study aims to evaluate the diagnostic performance of metagenomic third-generation sequencing (mTGS) using the Oxford Nanopore platform (Oxford Nanopore Technologies) for the detection of fungal pathogens in lower respiratory tract infections, and to compare its effectiveness with conventional diagnostic methods.
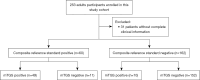
Methods: This study evaluated the clinical utility of mTGS with the Nanopore platform for diagnosing lower respiratory tract fungal infections (LRTFIs). Between January and August 2022, bronchoalveolar lavage fluid (BALF) samples were collected from 253 patients with suspected fungal infections across four medical centers in Hangzhou, China. Fungal detection was performed through both mTGS and conventional culture, and diagnostic performance was assessed via bioinformatics analysis and clinical adjudication.
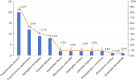
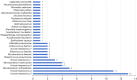
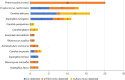
Results: Fungal infections were detected in 65 samples (29.3%), with 11 fungal species identified by mTGS, including Pneumocystis jirovecii, Cryptococcus neoformans, and Aspergillus fumigatus. Conventional culture identified only six species, missing key pathogens such as Pneumocystis jirovecii and Talaromyces marneffei. mTGS demonstrated a sensitivity of 78.1% [95% confidence interval (CI): 66.0-87.5%], a specificity of 90.5% (95% CI: 84.8-94.7%), a positive predictive value (PPV) of 76.9% (95% CI: 64.8-86.5%), and a negative predictive value (NPV) of 91.1% (95% CI: 85.4-95.0%). It showed high sensitivity for Pneumocystis jirovecii (76.5%) and Cryptococcus neoformans (82.4%) but lower sensitivity for Aspergillus spp. (66.7%). mTGS also co-detected viral and bacterial pathogens, offering comprehensive pathogen profiling, and significantly shortened the TATs to 7 hours as compared to 2-7 days for culture.
Conclusions: mTGS on the nanopore platform offers a rapid, sensitive, and comprehensive approach for diagnosing LRTFIs, particularly in immunocompromised patients. It serves as a valuable complementary tool for detecting mixed infections or culture-negative pneumonia. However, careful interpretation is needed regarding the clinical relevance of colonizing fungi such as Candida albicans.
Keywords: Metagenomic third-generation sequencing (mTGS); bronchoalveolar lavage fluid (BALF); lower respiratory tract fungal infections (LRTFIs); nanopore sequencing.
Copyright © 2025 AME Publishing Company. All rights reserved.
Conflict of interest statement
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jtd.amegroups.com/article/view/10.21037/jtd-2025-1163/coif). The authors have no conflicts of interest to declare.
Figures

References
LinkOut - more resources
Full Text Sources
Miscellaneous
