Application of machine learning in combination with mechanistic modeling to predict plasma exposure of small molecules
- PMID: 40809487
- PMCID: PMC12342024
- DOI: 10.3389/fsysb.2023.1180948
Application of machine learning in combination with mechanistic modeling to predict plasma exposure of small molecules
Abstract
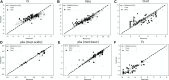
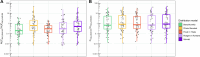
Prediction of a new molecule's exposure in plasma is a critical first step toward understanding its efficacy/toxicity profile and concluding whether it is a possible first-in-class, best-in-class candidate. For this prediction, traditional pharmacometrics use a variety of scaling methods that are heavily based on pre-clinical pharmacokinetic (PK) data. We here propose a novel framework based on which preclinical exposure prediction is performed by applying machine learning (ML) in tandem with mechanism-based modeling. In our proposed method, a relationship is initially established between molecular structure and physicochemical (PC)/PK properties using ML, and then the ML-driven PC/PK parameters are used as input to mechanistic models that ultimately predict the plasma exposure of new candidates. To understand the feasibility of our proposed framework, we evaluated a number of mechanistic models (1-compartment, physiologically based pharmacokinetic (PBPK)), PBPK distribution models (Berezhkovskiy, PK-Sim standard, Poulin and Theil, Rodgers and Rowland, and Schmidt), and PBPK parameterizations (using in vivo, or in vitro clearance). For most of the scenarios tested, our results demonstrate that PK profiles can be adequately predicted based on the proposed framework. Our analysis further indicates some limitations when liver microsomal intrinsic clearance (CLint) is used as the only clearance pathway and underscores the necessity of investigating the variability emanating from the different distribution models when providing PK predictions. The suggested approach aims at earlier exposure prediction in the drug development process so that critical decisions on molecule screening, chemistry design, or dose selection can be made as early as possible.
Keywords: PBPK; QSAR; artificial intelligence; drug discovery; machine learning; model-based drug development; pharmacokinetics.
Copyright © 2023 Mavroudis, Teutonico, Abos and Pillai.
Conflict of interest statement
All authors of this article are employees of Sanofi.
Figures




Similar articles
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
Comparison of Two Modern Survival Prediction Tools, SORG-MLA and METSSS, in Patients With Symptomatic Long-bone Metastases Who Underwent Local Treatment With Surgery Followed by Radiotherapy and With Radiotherapy Alone.Clin Orthop Relat Res. 2024 Dec 1;482(12):2193-2208. doi: 10.1097/CORR.0000000000003185. Epub 2024 Jul 23. Clin Orthop Relat Res. 2024. PMID: 39051924
-
Machine learning framework to predict pharmacokinetic profile of small molecule drugs based on chemical structure.Clin Transl Sci. 2024 May;17(5):e13824. doi: 10.1111/cts.13824. Clin Transl Sci. 2024. PMID: 38752574 Free PMC article.
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
-
Approaches for predicting dairy cattle methane emissions: from traditional methods to machine learning.J Anim Sci. 2024 Jan 3;102:skae219. doi: 10.1093/jas/skae219. J Anim Sci. 2024. PMID: 39123286 Free PMC article.
Cited by
-
The dawn of a new era: can machine learning and large language models reshape QSP modeling?J Pharmacokinet Pharmacodyn. 2025 Jun 16;52(4):36. doi: 10.1007/s10928-025-09984-5. J Pharmacokinet Pharmacodyn. 2025. PMID: 40524056 Free PMC article. Review.
References
-
- Antontsev V., Jagarapu A., Bundey Y., Hou H., Khotimchenko M., Walsh J., et al. (2021). A hybrid modeling approach for assessing mechanistic models of small molecule partitioning in vivo using a machine learning-integrated modeling platform. Sci. Rep. 11 (1), 11143. 10.1038/s41598-021-90637-1 - DOI - PMC - PubMed
-
- Awad M., Khanna R. (2015). “Support vector regression,” in Efficient learning machines: Theories, concepts, and applications for engineers and system designers (Berkeley, CA: Apress; ), 67–80.
LinkOut - more resources
Full Text Sources
