Phylogenetic Relationships and Evolutionary History of Goats (Mammalia: Capra) From Türkiye and Iraq, Inferred From Complete Mitochondrial Genomes
- PMID: 40809823
- PMCID: PMC12349903
- DOI: 10.1002/ece3.71985
Phylogenetic Relationships and Evolutionary History of Goats (Mammalia: Capra) From Türkiye and Iraq, Inferred From Complete Mitochondrial Genomes
Abstract
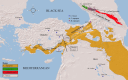
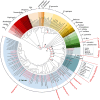
This study investigated genetic diversity, phylogenetic relationships, and evolutionary history of domestic goats from Türkiye and Iraq, along with wild goat and chamois species, using newly obtained mitogenomic sequences. Phylogenetic and phylogeographic analyses revealed a complex genetic structure among domestic goats, shaped by widespread distribution and gene flow. While haplotype A was predominant among domesticated breeds from both Türkiye and Iraq, haplotype G was also detected in the Turkish breeds. Notably, Turkish samples exhibited relatively higher nucleotide diversity (0.00133) compared to those from Iraq (0.00081), indicating greater genetic variability in the former population. Wild goat populations in Türkiye were clustered into two distinct lineages: (i) the Aegagrus lineage included the Artvin sample, some ancient genomes from the Taurus Mountains, and Iranian goats, and (ii) the Caucasian lineage contained Konya and Antalya samples, and some ancient genomes from the Taurus Mountains that were clustered closely with wild goats from the Caucasus. These findings suggest that geographic and ecological factors, such as the Anatolian Diagonal, influenced their diversification. Divergence time analyses indicated that the Caprinae began diversifying approximately 8.18 Mya, with initial splits in the Capra occurring around 3.22 Mya during the climatic fluctuations of the Late Pliocene/Early Pleistocene. The study also estimated the divergence of C. aegagrus and C. hircus at approximately 0.89 Mya in the Calabrian, with genetic diversification within domestic goats commencing 0.29 Mya in the Chibanian. The results provided robust evidence supporting Türkiye's role as a significant genetic center for goat domestication during the Neolithic period (~10,000 years ago). This hypothesis was further supported by the widespread presence of the common haplotype A in domestic goats, the high genetic diversity observed among domestic goats, and the region's proximity to the Fertile Crescent. The study underscored the importance of comprehensive genetic analyses in elucidating the evolutionary processes underlying goat domestication and highlighted the necessity for larger datasets and additional molecular markers to resolve the taxonomic complexities of wild goat populations in Türkiye, Iraq, and surrounding regions.
Keywords: Capra; Fertile Crescent; domestication; mitogenome; phylogeography; rupicapra.
© 2025 The Author(s). Ecology and Evolution published by British Ecological Society and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
Out of Africa: genetic characterization and diversity of Mashona cattle in the United States.J Anim Sci. 2025 Jan 4;103:skaf045. doi: 10.1093/jas/skaf045. J Anim Sci. 2025. PMID: 39951431
-
Does Augmenting Irradiated Autografts With Free Vascularized Fibula Graft in Patients With Bone Loss From a Malignant Tumor Achieve Union, Function, and Complication Rate Comparably to Patients Without Bone Loss and Augmentation When Reconstructing Intercalary Resections in the Lower Extremity?Clin Orthop Relat Res. 2025 Jun 26;483(9):1680-95. doi: 10.1097/CORR.0000000000003599. Online ahead of print. Clin Orthop Relat Res. 2025. PMID: 40569278
-
Global haplotype distribution of Babesia ovis inferred by 18S rRNA sequences; a phylogeographical systematic review.Microb Pathog. 2023 Aug;181:106179. doi: 10.1016/j.micpath.2023.106179. Epub 2023 May 26. Microb Pathog. 2023. PMID: 37245631
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
References
-
- Ağaoğlu, Ö. K. , and Ertuğrul O.. 2012. “Assessment of Genetic Diversity, Genetic Relationship and Bottleneck Using Microsatellites in Some Native Turkish Goat Breeds.” Small Ruminant Research 105, no. 1–3: 53–60. 10.1016/j.smallrumres.2011.12.005. - DOI
-
- Ahmadzadeh, F. , Flecks M., Rödder D., et al. 2013. “Multiple Dispersal out of Anatolia: Biogeography and Evolution of Oriental Green Lizards.” Biological Journal of the Linnean Society 110, no. 2: 398–408. 10.1111/bij.12129. - DOI
-
- Ansell, S. W. , Stenøien H. K., Grundmann M., et al. 2011. “The Importance of Anatolian Mountains as the Cradle of Global Diversity in <styled-content style="fixed-case"> Arabis alpina </styled-content>, a Key Arctic–Alpine Species.” Annals of Botany 108, no. 2: 241–252. 10.1093/aob/mcr134. - DOI - PMC - PubMed
-
- Balkız, O. 2002. “The Phylogeny of Wild Goats (<styled-content style="fixed-case"> Capra aegagrus </styled-content> E.) in Turkey as Determined by mtDNA D‐Loop Sequencing.” METU. Ankara. M. Sc. Thesis. 91 p.
LinkOut - more resources
Full Text Sources

