NewtCap: An Efficient Target Capture Approach to Boost Genomic Studies in Salamandridae (True Salamanders and Newts)
- PMID: 40809825
- PMCID: PMC12343749
- DOI: 10.1002/ece3.71835
NewtCap: An Efficient Target Capture Approach to Boost Genomic Studies in Salamandridae (True Salamanders and Newts)
Abstract
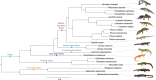
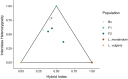
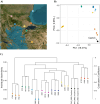
Salamanders have large and complex genomes, hampering whole genome sequencing. However, reduced representation sequencing provides a feasible alternative to obtain genome-wide data. We present NewtCap: a sequence capture bait set that targets c. 7 k coding regions across the genomes of all true salamanders and newts (the family Salamandridae, also known as "salamandrids"). We test the efficacy of NewtCap, originally designed for the Eurasian Triturus newts, in 30 species, belonging to 17 different genera that cover all main Salamandridae lineages. We also test NewtCap in two other salamander families. We discover that NewtCap performs well across all Salamandridae lineages (but not in the salamander families Ambystomatidae and Hynobiidae). As expected, the amount of genetic divergence from the genus Triturus correlates negatively to capture efficacy and mapping success. However, this does not impede our downstream analyses. We showcase the potential of NewtCap in the contexts of; (1) phylogenomics, by reconstructing the phylogeny of Salamandridae, (2) phylogeography, by sequencing the four closely related species comprising the genus Taricha, (3) hybrid zone analysis, by genotyping two Lissotriton species and different classes of interspecific hybrids, and (4) conservation genetics, by comparing Triturus ivanbureschi samples from several wild populations and one captive-bred population. Overall, NewtCap has the potential to boost straightforward, reproducible, and affordable genomic studies, tackling both fundamental and applied research questions across salamandrids.
Keywords: Caudata; Urodela; exon capture; high throughput sequencing; hyb‐seq; target enrichment.
© 2025 The Author(s). Ecology and Evolution published by British Ecological Society and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
Discordance between phylogenomic methods in Near Eastern mountain newts (Neurergus, Salamandridae).Mol Phylogenet Evol. 2025 Oct;211:108386. doi: 10.1016/j.ympev.2025.108386. Epub 2025 Jun 14. Mol Phylogenet Evol. 2025. PMID: 40523419
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
Identification of Y-chromosome turnover in newts fails to support a sex chromosome origin for the Triturus balanced lethal system.Genome Biol Evol. 2025 Aug 14:evaf155. doi: 10.1093/gbe/evaf155. Online ahead of print. Genome Biol Evol. 2025. PMID: 40810193
-
Omega-3 fatty acids for depression in adults.Cochrane Database Syst Rev. 2015 Nov 5;2015(11):CD004692. doi: 10.1002/14651858.CD004692.pub4. Cochrane Database Syst Rev. 2015. Update in: Cochrane Database Syst Rev. 2021 Nov 24;11:CD004692. doi: 10.1002/14651858.CD004692.pub5. PMID: 26537796 Free PMC article. Updated.
-
Antidepressants for pain management in adults with chronic pain: a network meta-analysis.Health Technol Assess. 2024 Oct;28(62):1-155. doi: 10.3310/MKRT2948. Health Technol Assess. 2024. PMID: 39367772 Free PMC article.
References
-
- Albert, T. J. , Molla M. N., Muzny D. M., et al. 2007. “Direct Selection of Human Genomic Loci by Microarray Hybridization.” Nature Methods 4: 903–905. - PubMed
-
- Alfaro, M. E. , Faircloth B. C., Harrington R. C., et al. 2018. “Explosive Diversification of Marine Fishes at the Cretaceous‐Palaeogene Boundary.” Nature Ecology & Evolution 2: 688–696. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

