Identification and validation of ubiquitination-related genes for predicting cervical cancer outcome
- PMID: 40809844
- PMCID: PMC12343280
- DOI: 10.3389/fgene.2025.1578075
Identification and validation of ubiquitination-related genes for predicting cervical cancer outcome
Abstract
Introduction: Abnormalities in ubiquitination-related pathways or systems are closely associated with various cancers, including cervical cancer (CC). However, the biological function and clinical value of ubiquitination-related genes (UbLGs) in CC remain unclear. This study aimed to explore key UbLGs associated with CC, construct a prognostic model, and investigate their potential clinical and immunological significance.
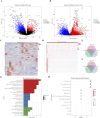
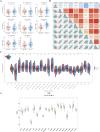
Methods: Differentially expressed genes (DEGs) between CC (tumor) and standard samples in self-sequencing and TCGA-GTEx-CESC datasets were identified using differential analysis. We identified overlaps between DEGs in both datasets and UbLGs, revealing key crossover genes. Subsequently, biological markers were identified via univariate Cox regression analysis and least absolute shrinkage and selection operator algorithms. After conducting independent prognostic analysis, immune infiltration analysis was performed to investigate the immune cells that differed between the two risk subgroups. Differences in immune checkpoint expression between the subgroups were analyzed. Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR) was performed to confirm the expression trends of the biomarkers.
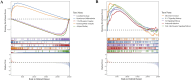
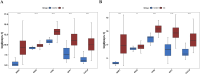
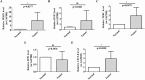
Results: Differentially expressed genes related to ubiquitination were screened from the Self-seq and TCGAGTEx-CESC datasets, and five key biomarkers (MMP1, RNF2, TFRC, SPP1, and CXCL8) were identified. The risk score model constructed based on these biomarkers could effectively predict the survival rate of cervical cancer patients (AUC >0.6 for 1/3/5 years). Immune microenvironment analysis showed that 12 types of immune cells, including memory B cells and M0 macrophages, as well as four immune checkpoints, exhibited significant differences between the high-risk and low-risk groups. RT-qPCR confirmed that MMP1, TFRC, and CXCL8 were upregulated in tumor tissues.
Discussion: Our study identified five ubiquitination-related biomarkers, namely, MMP1, RNF2, TFRC, SPP1, and CXCL8, which were significantly associated with CC. The validated risk model demonstrates strong predictive value for patient survival. These findings provide crucial insights into the role of ubiquitination in CC pathogenesis and offer valuable targets for advancing future research and therapeutic strategies.
Keywords: bioinformatics analysis; biomarker; cervical cancer; prognosis; ubiquitination.
Copyright © 2025 Jin, Fan, Liang, Dai and Wang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Allemani C., Matsuda T., Di Carlo V., Harewood R., Matz M., Nikšić M., et al. (2018). Global surveillance of trends in cancer survival 2000-14 (CONCORD-3): analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. Lancet 391 (10125), 1023–1075. 10.1016/S0140-6736(17)33326-3 - DOI - PMC - PubMed
-
- Bălăşoiu M., Bălăşoiu A. T., Mogoantă S., Bărbălan A., Stepan A. E., Ciurea R. N., et al. (2014). Serum and tumor microenvironment IL-8 values in different stages of colorectal cancer. Rom. J. Morphol. Embryol. 55 (2 Suppl. l), 575–578. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

