SETD6 mediates selective interaction and genomic occupancy of BRD4 and MITF in melanoma cells
- PMID: 40809945
- PMCID: PMC12342177
- DOI: 10.1093/narcan/zcaf023
SETD6 mediates selective interaction and genomic occupancy of BRD4 and MITF in melanoma cells
Abstract
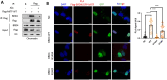
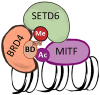
Aberrant transcriptional programs mediate malignant transformation of melanoma, the most aggressive form of skin cancer. The lysine methyltransferase SETD6 has been implicated in regulating transcription, cell adhesion, migration, and other processes in various cancers; however its role in melanoma remains unexplored. We recently reported that SETD6 monomethylates the BRD4 at K99 to selectively regulate transcription of genes involved in mRNA (messenger RNA) translation. Here, we observed that BRD4 methylation at K99 by SETD6 occurs in melanoma cells. Knockout of SETD6 or a point mutation at BRD4-K99 disrupts BRD4 genomic occupancy. In addition, we show that SETD6 interacts with MITF, a master transcription factor in melanocytes and melanoma, and influences the genomic distribution of MITF. Mechanistically, we uncover a novel chromatin-localized interaction between BRD4 and MITF in melanoma. Our data suggest that BRD4 binds MITF in melanoma cells and that this interaction is dependent on both SETD6-mediated methylation of BRD4 and MITF acetylation. This chromatin complex plays a pivotal role in selective recruitment of BRD4 and MITF to different genomic loci in melanoma cells.
© The Author(s) 2025. Published by Oxford University Press.
Conflict of interest statement
None declared.
Figures







References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

