Chloroplast Z-ring dynamics is governed by conserved core regions of evolutionarily divergent FtsZs
- PMID: 40810012
- PMCID: PMC12343686
- DOI: 10.3389/fpls.2025.1622675
Chloroplast Z-ring dynamics is governed by conserved core regions of evolutionarily divergent FtsZs
Abstract
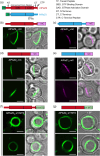
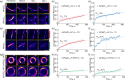
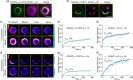
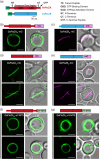
The chloroplast FtsZ ring (Z ring) is assembled by two distinct FtsZ proteins, FtsZ2 and FtsZ1 (referred to as FtsZA and FtsZB in red algae). FtsZ2 confers stability to the Z ring, while FtsZ1 enhances its dynamics. Enhanced Z-ring dynamics is essential for Z-ring remodeling, which drives chloroplast constriction and division. However, the mechanisms underlying the distinct dynamic properties of the two FtsZs remain unclear. Here, we report that the conserved core regions are primarily responsible for the distinct dynamic properties observed in both plant and red algal FtsZs. We demonstrate that the conserved core region of FtsZ1 enhances the dynamics of FtsZ2 within coassembled filaments. Likewise, we show that the conserved core region of red algal FtsZB promotes the dynamics of coassembled FtsZA rings. Our findings provide evidence that the evolution of a second FtsZ protein represents a general mechanism to enhance the dynamics of the chloroplast Z ring.
Keywords: FtsZ-ring; chloroplast; division; dynamics; evolution.
Copyright © 2025 Cao, Porter, Du, Tallerday, Liu, Liang, Osteryoung and Chen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Conserved Dynamics of Chloroplast Cytoskeletal FtsZ Proteins Across Photosynthetic Lineages.Plant Physiol. 2018 Jan;176(1):295-306. doi: 10.1104/pp.17.00558. Epub 2017 Aug 16. Plant Physiol. 2018. PMID: 28814573 Free PMC article.
-
Enhanced chloroplast FtsZ-ring constriction by the ARC6-ARC3 module in Arabidopsis.Proc Natl Acad Sci U S A. 2025 Jul;122(26):e2425129122. doi: 10.1073/pnas.2425129122. Epub 2025 Jun 25. Proc Natl Acad Sci U S A. 2025. PMID: 40560617
-
Evolution and Functional Differentiation of the C-terminal Motifs of FtsZs During Plant Evolution.Mol Biol Evol. 2024 Jul 3;41(7):msae145. doi: 10.1093/molbev/msae145. Mol Biol Evol. 2024. PMID: 39004892 Free PMC article.
-
Topical antiseptics for chronic suppurative otitis media.Cochrane Database Syst Rev. 2025 Jun 9;6(6):CD013055. doi: 10.1002/14651858.CD013055.pub3. Cochrane Database Syst Rev. 2025. PMID: 40484403
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
References
LinkOut - more resources
Full Text Sources

