The WD40 gene family in recretohalophyte Limonium bicolor: genomic identification and functional analysis in salt gland development and salinity tolerance
- PMID: 40810015
- PMCID: PMC12343514
- DOI: 10.3389/fpls.2025.1629604
The WD40 gene family in recretohalophyte Limonium bicolor: genomic identification and functional analysis in salt gland development and salinity tolerance
Abstract
Introduction: Developing salt-tolerant crops is critical for utilizing saline soils in agriculture. Limonium bicolor, a recretohalophyte with epidermal salt glands, represents a valuable genetic resource for salt tolerance engineering. Although WD40 proteins are known regulators of plant stress responses, their roles in L. bicolor remain unexplored.
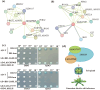
Methods: We performed a genome-wide analysis of WD40 genes in L. bicolor, including phylogenetic classification, subcellular localization prediction, cis-element analysis, and expression profiling during salt stress. Functional validation was conducted using virus-induced gene silencing (VIGS).
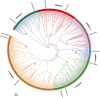
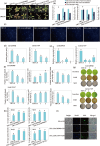
Results: Among 367 identified WD40 genes (distributed across all chromosomes), Subfamily 6 was the largest. Two key members (Lb1G05968 and Lb3G17197, localized in cytoplasm) showed significant involvement in salt gland development and stress tolerance, as demonstrated by VIGS-induced phenotypic defects.
Discussion: Our findings reveal the WD40 family's expansion in L. bicolor and its functional specialization in salt adaptation. The identified genes (e.g., Lb1G05968, Lb3G17197) provide targets for engineering salt-tolerant crops. This study establishes a foundation for further research on halophyte developmental genetics.
Keywords: Limonium bicolor; WD40 gene family; feature description; salt gland development; salt tolerance; subcellular localization.
Copyright © 2025 Sun, Mu, Tan, Wang, Wang and Yuan.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
The genome of the recretohalophyte Limonium bicolor provides insights into salt gland development and salinity adaptation during terrestrial evolution.Mol Plant. 2022 Jun 6;15(6):1024-1044. doi: 10.1016/j.molp.2022.04.011. Epub 2022 May 5. Mol Plant. 2022. PMID: 35514085
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
The WRKY gene family in the halophyte Limonium bicolor: identification, expression analysis, and regulation of salt stress tolerance.Plant Cell Rep. 2024 Jun 12;43(7):167. doi: 10.1007/s00299-024-03258-z. Plant Cell Rep. 2024. PMID: 38865016
-
Learning from Salicornia: Physiological, Biochemical, and Molecular Mechanisms of Salinity Tolerance.Int J Mol Sci. 2025 Jun 20;26(13):5936. doi: 10.3390/ijms26135936. Int J Mol Sci. 2025. PMID: 40649711 Free PMC article. Review.
-
Diagnostic test accuracy and cost-effectiveness of tests for codeletion of chromosomal arms 1p and 19q in people with glioma.Cochrane Database Syst Rev. 2022 Mar 2;3(3):CD013387. doi: 10.1002/14651858.CD013387.pub2. Cochrane Database Syst Rev. 2022. PMID: 35233774 Free PMC article.
References
-
- Awdhesh K. M., Puranik S., Prasad M. (2012). Structure and regulatory networks of WD40 protein in plants. J. Plant Biochem. Biotechnol. 21, 32–39. doi: 10.1007/s13562-012-0134-1 - DOI
-
- Carey C. C., Strahle J. T., Selinger D. A., Chandler V. L. (2004). Mutations in the pale aleurone color1 regulatory gene of the Zea mays anthocyanin pathway have distinct phenotypes relative to the functionally similar TRANSPARENT TESTA GLABRA1 gene in Arabidopsis thaliana. Plant Cell 16, 450–464. doi: 10.1105/tpc.018796, PMID: - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

