A tubulin-MAPKKK pathway engages tubulin isotype interaction for neuroprotection
- PMID: 40811477
- PMCID: PMC12403078
- DOI: 10.1073/pnas.2507208122
A tubulin-MAPKKK pathway engages tubulin isotype interaction for neuroprotection
Abstract
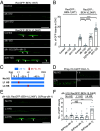
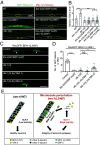
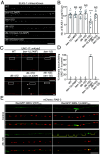
The microtubule (MT) cytoskeleton is essential for neuronal morphology, neurite growth, synapse formation and maintenance, as well as regulation of signal transduction. Most cells express multiple isotypes of α- and β-tubulin that can coassemble into MTs. While a variety of signaling pathways regulate MT integrity and homeostasis, little is known about how tubulin isotypes interact in vivo. Here, we report a mechanism in which altered function of a neuronal β-tubulin in Caenorhabditis elegans activates the conserved kinase DLK-1 and its downstream signal transduction, which in turn upregulates expression of an α-tubulin isotype to ensure MT integrity. We find that alteration in the T7 loop of the β-tubulin/BEN-1 causes the formation of BEN-1-enriched islands along MTs in neurites. Combining genome editing with cellular imaging, we identified amino acid residues in α-tubulin/TBA-2 that are necessary for formation of BEN-1 islands. Activation of DLK-1 signaling in ben-1 mutants promotes TBA-2 transcription and protects axon and synapse morphology. These data uncover a positive feedback loop between DLK-1 and regulation of tubulin isotype interaction that maintains neuronal resilience.
Keywords: DLK kinase; bZip protein CEBP-1; microtubule plasticity; neuronal resilience; tubulin isotype.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures




References
-
- Kapitein L. C., Hoogenraad C. C., Building the neuronal microtubule cytoskeleton. Neuron 87, 492–506 (2015). - PubMed
-
- Cason S. E., Holzbaur E. L. F., Selective motor activation in organelle transport along axons. Nat. Rev. Mol. Cell Biol. 23, 699–714 (2022). - PubMed
-
- Dumont E. L., Do C., Hess H., Molecular wear of microtubules propelled by surface-adhered kinesins. Nat. Nanotechnol. 10, 166–169 (2015). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

