Chromatin retained MUSHER lncRNA integrates ABA and DOG1 signalling pathways to enhance Arabidopsis seeds dormancy
- PMID: 40813580
- PMCID: PMC12354759
- DOI: 10.1038/s41467-025-62991-5
Chromatin retained MUSHER lncRNA integrates ABA and DOG1 signalling pathways to enhance Arabidopsis seeds dormancy
Abstract
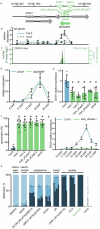
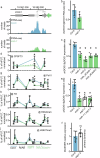
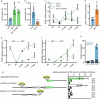
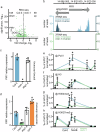
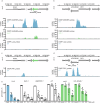
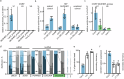
Many plant lncRNAs regulate gene expression by binding to chromatin, but how they are retained at the target loci is unclear. We identify a chromatin-localized lncRNA - MUSHER, which activates two parallel regulatory pathways to increase Arabidopsis seed dormancy. MUSHER is upregulated in response to high temperatures, contributing to the induction of secondary dormancy. It promotes DOG1 expression by recruitment of the CPSF complex to enhance the proximal cleavage and polyadenylation at the DOG1 transcript. It also increases ABA sensitivity in seeds by activating PIR1 gene transcription. These genes, located on different chromosomes, are both bound by MUSHER, despite lacking sequence homology. The chromatin association of MUSHER enables the integration of the DOG1- and ABA pathways to adjust seed germination timing. Additionally, MUSHER and other lncRNAs interact with U1 snRNP, which is required for their chromatin localisation, revealing a role for U1 snRNP in plants.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: K.J.R. and S.S. have equity interests in ElementZero Biolabs UG. S.S. also serve on the Scientific Advisory Board for ElementZero Biolabs UG. All other authors declare no competing interests.
Figures

References
-
- Ariel, F. et al. R-loop mediated trans action of the APOLO long noncoding RNA. Mol. Cell77, 1055–1065.e4 (2020). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

