Multimodal spatial transcriptomic characterization of mouse kidney injury and repair
- PMID: 40813851
- PMCID: PMC12354736
- DOI: 10.1038/s41467-025-62599-9
Multimodal spatial transcriptomic characterization of mouse kidney injury and repair
Abstract
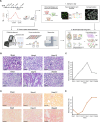
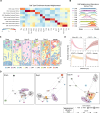
The transition from acute kidney injury to chronic kidney disease is characterized by significant changes in the cellular composition and molecular interactions within the kidney. Utilizing high-resolution Xenium and whole transcriptome Visium spatial transcriptomics platforms, we analyze over a million cells on 12 male mouse kidneys across six stages of renal injury and repair. We define and validate 20 major kidney cell populations and delineate distinct cellular neighborhoods through this multimodal spatial analysis. We further reveal a specific fibro-inflammatory niche enriched in failed-repair proximal tubule cells, fibroblasts, and immune cells, with conserved neighborhood gene signatures across mouse and human. Within this niche, we predict Runx2 as a key upstream regulator, along with platelet-derived growth factor and integrin beta-2 signaling pathways shaping the fibrogenic microenvironment. Altogether, our study provides deep insights into the cellular and molecular dynamics during kidney injury and repair and establishes a comprehensive multimodal analytical framework applicable to other spatial omics studies.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




References
-
- Bellomo, R., Kellum, J. A. & Ronco, C. Acute kidney injury. Lancet380, 756–766 (2012). - PubMed
-
- Jager, K. J. et al. A single number for advocacy and communication—worldwide more than 850 million individuals have kidney diseases. Nephrol. Dial. Transpl.34, 1803–1805 (2019). - PubMed
-
- Kumar, S. Cellular and molecular pathways of renal repair after acute kidney injury. Kidney Int93, 27–40 (2018). - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

