Neoadjuvant Immunotherapy Promotes the Formation of Mature Tertiary Lymphoid Structures in a Remodeled Pancreatic Tumor Microenvironment
- PMID: 40815230
- PMCID: PMC12424053
- DOI: 10.1158/2326-6066.CIR-25-0387
Neoadjuvant Immunotherapy Promotes the Formation of Mature Tertiary Lymphoid Structures in a Remodeled Pancreatic Tumor Microenvironment
Abstract
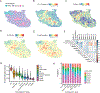
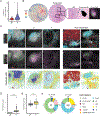
Pancreatic ductal adenocarcinoma (PDAC) is a rapidly progressing cancer that responds poorly to immunotherapies. Intratumoral tertiary lymphoid structures (TLS) have been associated with rare long-term PDAC survivors, but the role of TLS in PDAC and their spatial relationships within the context of the broader tumor microenvironment remain unknown. In this study, we report the generation of a spatial multiomic atlas of PDAC tumors and tumor-adjacent lymph nodes from patients treated with combination neoadjuvant immunotherapies. Using machine learning-enabled hematoxylin and eosin image classification models, imaging mass cytometry, and unsupervised gene expression matrix factorization methods for spatial transcriptomics, we characterized cellular states within and adjacent to TLS spanning distinct spatial niches and pathologic responses. Unsupervised learning identified TLS-specific spatial gene expression signatures that are significantly associated with improved survival in patients with PDAC. We identified spatial features of pathologic immune responses, including intratumoral TLS-associated B-cell maturation colocalizing with IgG dissemination and extracellular matrix remodeling. Our findings offer insights into the cellular and molecular landscape of TLS in PDACs during immunotherapy treatment.
©2025 American Association for Cancer Research.
Figures




References
-
- Timmer FEF, Geboers B, Nieuwenhuizen S, Dijkstra M, Schouten EAC, Puijk RS, et al. Pancreatic Cancer and Immunotherapy: A Clinical Overview. Cancers (Basel) 2021;13
MeSH terms
Grants and funding
- P50 CA062924/CA/NCI NIH HHS/United States
- R50 CA243627/CA/NCI NIH HHS/United States
- K00 AG068527/AG/NIA NIH HHS/United States
- P01 CA247886/CA/NCI NIH HHS/United States
- P01 CA114046/CA/NCI NIH HHS/United States
- F31 CA268724/CA/NCI NIH HHS/United States
- U01 CA253403/CA/NCI NIH HHS/United States
- U01 CA212007/CA/NCI NIH HHS/United States
- Lustgarten Foundation (Lustgarten)
- Sol Goldman Charitable Trustl
- NIH P01-CA247886-01A1/National Cancer Institute (NCI)
- SPORE GI P50CA062924-24A1/National Cancer Institute (NCI)
- Emerson Collective (Emerson)
- SU2C/AACR DT-14-14/American Association for Cancer Research (AACR)
- Allegheny Health Network (AHN)
- NIH U01CA212007/National Cancer Institute (NCI)
- NIH U01CA253403/National Cancer Institute (NCI)
- NCI R50 CA243627/National Cancer Institute (NCI)
- NCI F31CA268724-01/National Cancer Institute (NCI)
- Sanofi (Sanofi US)
- NIH K00AG068527 (NIA)/National Institute on Aging (NIA)
- Samuel Waxman Cancer Research Foundation (SWCRF)
- Mark Foundation For Cancer Research (The Mark Foundation for Cancer Research)
- NIH P01CA114046 (NCI)/National Cancer Institute (NCI)
- Discovery Award/Office of the Provost, Johns Hopkins University
LinkOut - more resources
Full Text Sources
Medical

