TRIM24 as a therapeutic target in endocrine treatment-resistant breast cancer
- PMID: 40815626
- PMCID: PMC12377727
- DOI: 10.1073/pnas.2507571122
TRIM24 as a therapeutic target in endocrine treatment-resistant breast cancer
Abstract
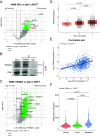
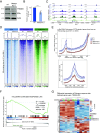
While Estrogen receptor alpha (ERα)+ breast cancer treatment is considered effective, resistance to endocrine therapy is common. Since ERα is still the main driver in most therapy-resistant tumors, alternative therapeutic strategies are needed to disrupt ERα transcriptional activity. In this work, we position TRIM24 as a therapeutic target in endocrine resistance, given its role as a key component of the ERα transcriptional complex. TRIM24 interacts with ERα and other well-known ERα cofactors to facilitate ERα chromatin interactions and allows for maintenance of active histone marks including H3K23ac and H3K27ac. Consequently, genetic perturbation of TRIM24 abrogates ERα-driven transcriptional programs and reduces tumor cell proliferation capacity. Using a recently developed degrader targeting TRIM24, ERα-driven transcriptional output and growth were blocked, effectively treating not only endocrine-responsive cell lines but also drug-resistant derivatives thereof as well as cell line models bearing activating ESR1 point mutations. Finally, using human tumor-derived organoid models, we could show the efficacy of TRIM24 degrader in the endocrine-responsive and -resistant setting. Overall, our study positions TRIM24 as a central component for the integrity and activity of the ERα transcriptional complex, with degradation-mediated perturbation of TRIM24 as a promising therapeutic avenue in the treatment of primary and endocrine resistance breast cancer.
Keywords: TRIM24; breast cancer; estrogen receptor alpha; heterobifunctional protein degrader; therapy resistance breast cancer.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

